4G36
 
 | |
4G37
 
 | | Structure of cross-linked firefly luciferase in second catalytic conformation | | Descriptor: | 4,4'-(ethylenediimino)bis[4-oxobutyrate], 5'-O-[N-(DEHYDROLUCIFERYL)-SULFAMOYL] ADENOSINE, CHLORIDE ION, ... | | Authors: | Sundlov, J.A, Branchini, B.R, Gulick, A.M. | | Deposit date: | 2012-07-13 | | Release date: | 2012-08-15 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.396 Å) | | Cite: | Crystal structure of firefly luciferase in a second catalytic conformation supports a domain alternation mechanism.
Biochemistry, 51, 2012
|
|
2RGN
 
 | | Crystal Structure of p63RhoGEF complex with Galpha-q and RhoA | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Guanine nucleotide-binding protein G(i) subunit alpha-1,Guanine nucleotide-binding protein G(q) subunit alpha, MAGNESIUM ION, ... | | Authors: | Shankaranarayanan, A, Nance, M.R, Tesmer, J.J.G. | | Deposit date: | 2007-10-04 | | Release date: | 2008-01-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of Galphaq-p63RhoGEF-RhoA complex reveals a pathway for the activation of RhoA by GPCRs.
Science, 318, 2007
|
|
4GZL
 
 | | Crystal structure of RAC1 Q61L mutant | | Descriptor: | ISOPROPYL ALCOHOL, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Ha, B.H, Boggon, T.J. | | Deposit date: | 2012-09-06 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | RAC1P29S is a spontaneously activating cancer-associated GTPase.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3LXR
 
 | | Shigella IpgB2 in complex with human RhoA and GDP (complex C) | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, IpgB2, SULFATE ION, ... | | Authors: | Klink, B.U, Barden, S, Heidler, T.V, Borchers, C, Ladwein, M, Stradal, T.E.B, Rottner, K, Heinz, D.W. | | Deposit date: | 2010-02-25 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structure of Shigella IPGB2 in complex with human RhoA: Implications for the mechanism of bacterial GEF-mimicry
J.Biol.Chem., 285, 2010
|
|
3LWN
 
 | | Shigella IpgB2 in complex with human RhoA, GDP and Mg2+ (complex B) | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, IpgB2, MAGNESIUM ION, ... | | Authors: | Klink, B.U, Barden, S, Heidler, T.V, Borchers, C, Ladwein, M, Stradal, T.E.B, Rottner, K, Heinz, D.W. | | Deposit date: | 2010-02-24 | | Release date: | 2010-03-31 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.28 Å) | | Cite: | Structure of Shigella IPGB2 in complex with human RhoA: Implications for the mechanism of bacterial GEF-mimicry
J.Biol.Chem., 285, 2010
|
|
7KQZ
 
 | |
3RYT
 
 | | The Plexin A1 intracellular region in complex with Rac1 | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Plexin-A1, ... | | Authors: | Zhang, X, He, H. | | Deposit date: | 2011-05-11 | | Release date: | 2012-01-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (3.582 Å) | | Cite: | Plexins Are GTPase-Activating Proteins for Rap and Are Activated by Induced Dimerization.
Sci.Signal., 5, 2012
|
|
8Q0N
 
 | | HACE1 in complex with RAC1 Q61L | | Descriptor: | E3 ubiquitin-protein ligase HACE1, GUANOSINE-5'-TRIPHOSPHATE, Ras-related C3 botulinum toxin substrate 1, ... | | Authors: | Wolter, M, Duering, J, Dienemann, C, Lorenz, S. | | Deposit date: | 2023-07-28 | | Release date: | 2024-01-10 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural mechanisms of autoinhibition and substrate recognition by the ubiquitin ligase HACE1.
Nat.Struct.Mol.Biol., 31, 2024
|
|
3O83
 
 | | Structure of BasE N-terminal domain from Acinetobacter baumannii bound to 2-(4-n-dodecyl-1,2,3-triazol-1-yl)-5'-O-[N-(2-hydroxybenzoyl)sulfamoyl]adenosine | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 2-(4-dodecyl-1H-1,2,3-triazol-1-yl)-5'-O-{[(2-hydroxyphenyl)carbonyl]sulfamoyl}adenosine, ... | | Authors: | Drake, E.J, Duckworth, B.P, Neres, J, Aldrich, C.C, Gulick, A.M. | | Deposit date: | 2010-08-02 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Biochemical and structural characterization of bisubstrate inhibitors of BasE, the self-standing nonribosomal peptide synthetase adenylate-forming enzyme of acinetobactin synthesis.
Biochemistry, 49, 2010
|
|
3SBD
 
 | | Crystal structure of RAC1 P29S mutant | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Ha, B.H, Boggon, T.J. | | Deposit date: | 2011-06-03 | | Release date: | 2012-07-18 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Exome sequencing identifies recurrent somatic RAC1 mutations in melanoma.
Nat.Genet., 44, 2012
|
|
4FL2
 
 | | Structural and Biophysical Characterization of the Syk Activation Switch | | Descriptor: | MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, Tyrosine-protein kinase SYK | | Authors: | Graedler, U, Schwarz, D, Dresing, V, Musil, M, Bomke, J, Frech, M, Jaekel, S, Rysiok, T, Mueller-Pompalla, D, Wegener, A. | | Deposit date: | 2012-06-14 | | Release date: | 2012-11-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural and biophysical characterization of the syk activation switch.
J.Mol.Biol., 425, 2013
|
|
8FBP
 
 | | Glutamine synthetase from Pseudomonas aeruginosa, filament double-unit in compressed conformation | | Descriptor: | Glutamine synthetase | | Authors: | Phan, I.Q, Staker, B, Shek, R, Moser, T.H, Evans, J.E, van Voorhis, W.C, Myler, P.J, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-11-29 | | Release date: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Glutamine synthetase from Pseudomonas aeruginosa, filament double-unit in compressed conformation
To Be Published
|
|
4GR4
 
 | | Crystal structure of SlgN1deltaAsub | | Descriptor: | CHLORIDE ION, Non-ribosomal peptide synthetase | | Authors: | Herbst, D.A, Zocher, G, Stehle, T. | | Deposit date: | 2012-08-24 | | Release date: | 2012-11-28 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Structural Basis of the Interaction of MbtH-like Proteins, Putative Regulators of Nonribosomal Peptide Biosynthesis, with Adenylating Enzymes.
J.Biol.Chem., 288, 2013
|
|
4GZM
 
 | | Crystal structure of RAC1 F28L mutant | | Descriptor: | 5'-GUANOSINE-DIPHOSPHATE-MONOTHIOPHOSPHATE, MAGNESIUM ION, Ras-related C3 botulinum toxin substrate 1 | | Authors: | Ha, B.H, Boggon, T.J. | | Deposit date: | 2012-09-06 | | Release date: | 2012-12-12 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | RAC1P29S is a spontaneously activating cancer-associated GTPase.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3REU
 
 | | Crystal structure of the archaeal asparagine synthetase A complexed with L-Aspartic acid and Adenosine triphosphate | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, ASPARTIC ACID, AsnS-like asparaginyl-tRNA synthetase related protein, ... | | Authors: | Blaise, M, Frechin, M, Charron, C, Roy, H, Sauter, C, Lorber, B, Olieric, V, Kern, D. | | Deposit date: | 2011-04-05 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Archaeal Asparagine Synthetase: Interrelation with Aspartyl-tRNA and Asparaginyl-tRNA Synthetases.
J.Mol.Biol., 412, 2011
|
|
3R44
 
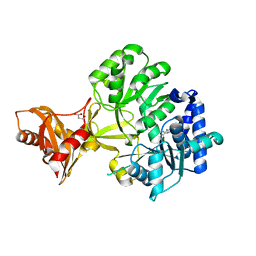 | | Mycobacterium tuberculosis fatty acyl CoA synthetase | | Descriptor: | HISTIDINE, MALONATE ION, fatty acyl CoA synthetase FADD13 (FATTY-ACYL-CoA SYNTHETASE) | | Authors: | Andersson, C.S, Martinez Molina, D, Hogbom, M. | | Deposit date: | 2011-03-17 | | Release date: | 2012-02-08 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The Mycobacterium tuberculosis Very-Long-Chain Fatty Acyl-CoA Synthetase: Structural Basis for Housing Lipid Substrates Longer than the Enzyme.
Structure, 20, 2012
|
|
3O84
 
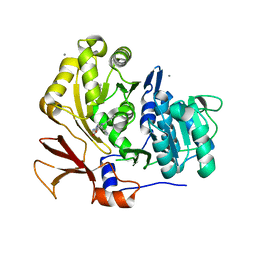 | | Structure of BasE N-terminal domain from Acinetobacter baumannii bound to 6-phenyl-1-(pyridin-4-ylmethyl)-1H-pyrazolo[3,4-b]pyridine-4-carboxylic acid. | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, 6-phenyl-1-(pyridin-4-ylmethyl)-1H-pyrazolo[3,4-b]pyridine-4-carboxylic acid, ... | | Authors: | Drake, E.J, Duckworth, B.P, Neres, J, Aldrich, C.C, Gulick, A.M. | | Deposit date: | 2010-08-02 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Biochemical and structural characterization of bisubstrate inhibitors of BasE, the self-standing nonribosomal peptide synthetase adenylate-forming enzyme of acinetobactin synthesis.
Biochemistry, 49, 2010
|
|
3O82
 
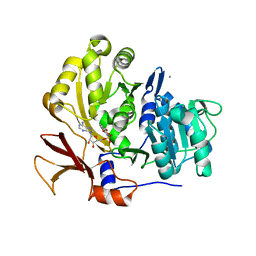 | | Structure of BasE N-terminal domain from Acinetobacter baumannii bound to 5'-O-[N-(2,3-dihydroxybenzoyl)sulfamoyl] adenosine | | Descriptor: | 5'-O-{[(2,3-dihydroxyphenyl)carbonyl]sulfamoyl}adenosine, CALCIUM ION, Peptide arylation enzyme | | Authors: | Drake, E.J, Duckworth, B.P, Neres, J, Aldrich, C.C, Gulick, A.M. | | Deposit date: | 2010-08-02 | | Release date: | 2010-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Biochemical and structural characterization of bisubstrate inhibitors of BasE, the self-standing nonribosomal peptide synthetase adenylate-forming enzyme of acinetobactin synthesis.
Biochemistry, 49, 2010
|
|
2VSQ
 
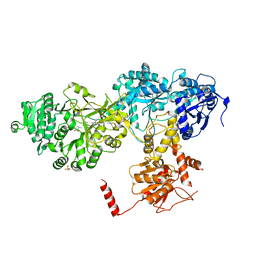 | | Structure of surfactin A synthetase C (SrfA-C), a nonribosomal peptide synthetase termination module | | Descriptor: | LEUCINE, SULFATE ION, SURFACTIN SYNTHETASE SUBUNIT 3 | | Authors: | Tanovic, A, Samel, S.A, Essen, L.-O, Marahiel, M.A. | | Deposit date: | 2008-04-29 | | Release date: | 2008-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the termination module of a nonribosomal peptide synthetase.
Science, 321, 2008
|
|
2VRW
 
 | |
8GJW
 
 | | Structure of a cGAS-like receptor Cv-cGLR1 from C. virginica | | Descriptor: | SULFATE ION, cGAS-like receptor 1 | | Authors: | Li, Y, Morehouse, B.R, Slavik, K.M, Liu, J, Toyoda, H, Kranzusch, P.J. | | Deposit date: | 2023-03-16 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | cGLRs are a diverse family of pattern recognition receptors in innate immunity.
Cell, 186, 2023
|
|
8GJX
 
 | | Structure of the human STING receptor bound to 2'3'-cUA | | Descriptor: | 2'3'-cUA, Stimulator of interferon genes protein | | Authors: | Morehouse, B.R, Li, Y, Slavik, K.M, Toyoda, H, Kranzusch, P.J. | | Deposit date: | 2023-03-16 | | Release date: | 2023-07-05 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | cGLRs are a diverse family of pattern recognition receptors in innate immunity.
Cell, 186, 2023
|
|
8GJZ
 
 | | Structure of a STING receptor from S. pistillata Sp-STING1 bound to 2'3'-cUA | | Descriptor: | 2'3'-cUA, Stimulator of interferon genes protein | | Authors: | Li, Y, Toyoda, H, Slavik, K.M, Morehouse, B.R, Kranzusch, P.J. | | Deposit date: | 2023-03-16 | | Release date: | 2023-07-05 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | cGLRs are a diverse family of pattern recognition receptors in innate immunity.
Cell, 186, 2023
|
|
2WMN
 
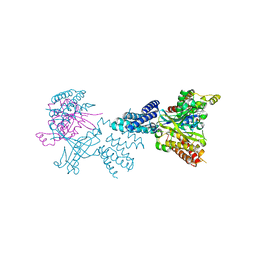 | | Structure of the complex between DOCK9 and Cdc42-GDP. | | Descriptor: | CELL DIVISION CONTROL PROTEIN 42 HOMOLOG, DEDICATOR OF CYTOKINESIS PROTEIN 9, GUANOSINE-5'-DIPHOSPHATE | | Authors: | Yang, J, Roe, S.M, Barford, D. | | Deposit date: | 2009-07-02 | | Release date: | 2009-09-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.391 Å) | | Cite: | Activation of Rho Gtpases by Dock Exchange Factors is Mediated by a Nucleotide Sensor.
Science, 325, 2009
|
|
