5LV0
 
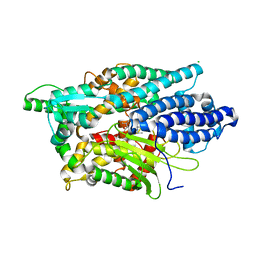 | | Structure of Human Neurolysin (E475Q) in complex with amyloid-beta 35-40 peptide product | | Descriptor: | CHLORIDE ION, GLY-VAL-VAL amyloid 35-40 fragment, Neurolysin, ... | | Authors: | Masuyer, G, Berntsson, R.P.-A, Teixeira, P.F, Kmiec, B, Glaser, E, Stenmark, P. | | Deposit date: | 2016-09-12 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and functional analysis of Neurolysin, a new component of the mitochondrial peptidolytic network
To Be Published
|
|
5LTS
 
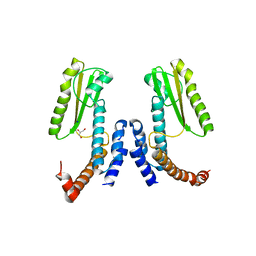 | | Crystal structure of Lymphocytic choriomeningitis mammarenavirus endonuclease Mutant D118A | | Descriptor: | CHLORIDE ION, GLYCEROL, RNA-directed RNA polymerase L, ... | | Authors: | Saez-Ayala, M, Yekwa, E.L, Canard, B, Alvarez, K, Ferron, F. | | Deposit date: | 2016-09-07 | | Release date: | 2017-09-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Crystal structures ofLymphocytic choriomeningitis virusendonuclease domain complexed with diketo-acid ligands.
IUCrJ, 5, 2018
|
|
8UFC
 
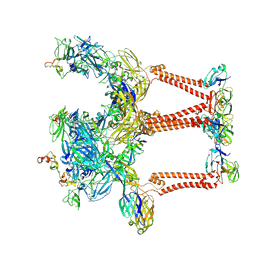 | |
5KBN
 
 | | The crystal structure of fluoride channel Fluc Ec2 F80I Mutant | | Descriptor: | DECYL-BETA-D-MALTOPYRANOSIDE, FLUORIDE ION, Putative fluoride ion transporter CrcB, ... | | Authors: | Last, N.B, Kolmakova-Partensky, L, Shane, T, Miller, C. | | Deposit date: | 2016-06-03 | | Release date: | 2016-08-03 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Mechanistic signs of double-barreled structure in a fluoride ion channel.
Elife, 5, 2016
|
|
8UFA
 
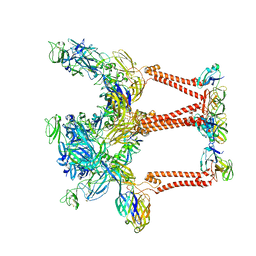 | |
1CDT
 
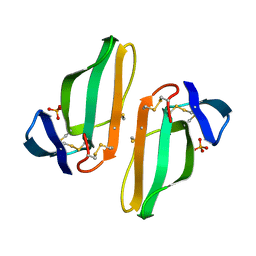 | | CARDIOTOXIN V4/II FROM NAJA MOSSAMBICA MOSSAMBICA: THE REFINED CRYSTAL STRUCTURE | | Descriptor: | CARDIOTOXIN VII4, PHOSPHATE ION | | Authors: | Rees, B, Bilwes, A, Samama, J.P, Moras, D. | | Deposit date: | 1990-05-17 | | Release date: | 1991-07-15 | | Last modified: | 2017-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Cardiotoxin VII4 from Naja mossambica mossambica. The refined crystal structure.
J.Mol.Biol., 214, 1990
|
|
1C6V
 
 | | SIV INTEGRASE (CATALYTIC DOMAIN + DNA BIDING DOMAIN COMPRISING RESIDUES 50-293) MUTANT WITH PHE 185 REPLACED BY HIS (F185H) | | Descriptor: | PROTEIN (SIU89134), PROTEIN (SIV INTEGRASE) | | Authors: | Chen, Z, Yan, Y, Munshi, S, Li, Y, Zruygay-Murphy, J, Xu, B, Witmer, M, Felock, P, Wolfe, A, Sardana, V, Emini, E.A, Hazuda, D, Kuo, L.C. | | Deposit date: | 1999-12-21 | | Release date: | 2000-12-27 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-ray structure of simian immunodeficiency virus integrase containing the core and C-terminal domain (residues 50-293)--an initial glance of the viral DNA binding platform.
J.Mol.Biol., 296, 2000
|
|
6FS9
 
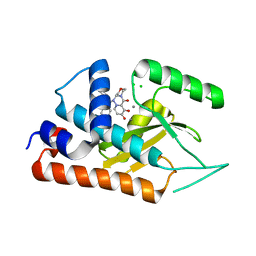 | |
7LWQ
 
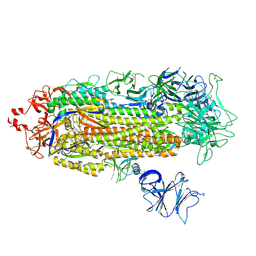 | |
8IUX
 
 | |
6HPQ
 
 | | Crystal structure of human Pif1 helicase in complex with AMP-PNP, brominated crystal form. | | Descriptor: | ATP-dependent DNA helicase PIF1, BROMIDE ION, MAGNESIUM ION, ... | | Authors: | Ledikov, V.M, Dehghani-Tafti, S, Bax, B, Sanders, C.M, Antson, A.A. | | Deposit date: | 2018-09-21 | | Release date: | 2019-01-23 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Structural and functional analysis of the nucleotide and DNA binding activities of the human PIF1 helicase.
Nucleic Acids Res., 47, 2019
|
|
8IV0
 
 | | H7N9 HA-C4H4 Fab | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, C4H4 Fab heavy chain, C4H4 Fab light chain, ... | | Authors: | Zhao, B.B, Sun, Z.Z. | | Deposit date: | 2023-03-25 | | Release date: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | H7N9 HA-C4H4 Fab
To Be Published
|
|
5M11
 
 | | Structural and functional probing of PorZ, an essential bacterial surface component of the type-IX secretion system of human oral-microbiomic Porphyromonas gingivalis. | | Descriptor: | CACODYLATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Lasica, A.M, Goulas, T, Mizgalska, D, Zhou, X, Ksiazek, M, Madej, M, Guo, Y, Guevara, T, Nowak, M, Potempa, B, Goel, A, Sztukowska, M, Prabhakar, A.T, Bzowska, M, Widziolek, M, Thogersen, I.B, Enghild, J.J, Simonian, M, Kulczyk, A.W, Nguyen, K.-A, Potempa, J, Gomis-Ruth, F.X. | | Deposit date: | 2016-10-07 | | Release date: | 2016-11-09 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural and functional probing of PorZ, an essential bacterial surface component of the type-IX secretion system of human oral-microbiomic Porphyromonas gingivalis.
Sci Rep, 6, 2016
|
|
7BM8
 
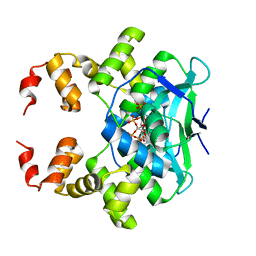 | | Crystal structure of the C-terminally truncated chromosome-partitioning protein ParB from Caulobacter crescentus complexed with CTP-gamma-S | | Descriptor: | CYTIDINE-5'-TRIPHOSPHATE, Chromosome-partitioning protein ParB, MAGNESIUM ION | | Authors: | Jalal, A.S, Tran, N.T, Stevenson, C.E.M, Lawson, D.M, Le, T.B.K. | | Deposit date: | 2021-01-19 | | Release date: | 2021-04-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.73 Å) | | Cite: | A CTP-dependent gating mechanism enables ParB spreading on DNA.
Elife, 10, 2021
|
|
5CL7
 
 | |
6T3G
 
 | |
5IER
 
 | |
6T3W
 
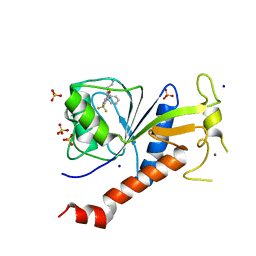 | | Coxsackie B3 2C protein in complex with S-Fluoxetine | | Descriptor: | (3S)-N-methyl-3-phenyl-3-[4-(trifluoromethyl)phenoxy]propan-1-amine, 2C protein, SODIUM ION, ... | | Authors: | El Kazzi, P, Papageorgiou, N, Ferron, F.P, Bauer, L, van Kuppeveld, F, Coutard, B. | | Deposit date: | 2019-10-11 | | Release date: | 2020-11-18 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.82 Å) | | Cite: | Fluoxetine targets an allosteric site in the enterovirus 2C AAA+ ATPase and stabilizes a ring-shaped hexameric complex.
Sci Adv, 8, 2022
|
|
6BMC
 
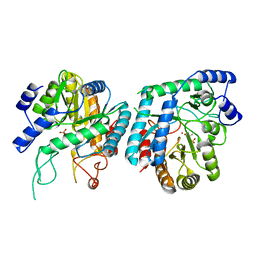 | | The structure of a dimeric type II DAH7PS associated with pyocyanin biosynthesis in Pseudomonas aeruginosa | | Descriptor: | CHLORIDE ION, COBALT (II) ION, PHOSPHOENOLPYRUVATE, ... | | Authors: | Sterritt, O.W, Jameson, G.B, Parker, E.J. | | Deposit date: | 2017-11-14 | | Release date: | 2018-10-03 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural and functional characterisation of the entry point to pyocyanin biosynthesis inPseudomonas aeruginosadefines a new 3-deoxy-d-arabino-heptulosonate 7-phosphate synthase subclass.
Biosci. Rep., 38, 2018
|
|
5EYL
 
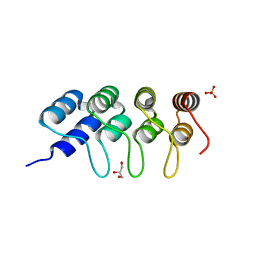 | | TUBULIN-BINDING DARPIN | | Descriptor: | DESIGNED ANKYRIN REPEAT PROTEIN (DARPIN), GLYCEROL, PHOSPHATE ION | | Authors: | Ahmad, S, Kossow, M, Gigant, B. | | Deposit date: | 2015-11-25 | | Release date: | 2016-07-20 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Destabilizing an interacting motif strengthens the association of a designed ankyrin repeat protein with tubulin.
Sci Rep, 6, 2016
|
|
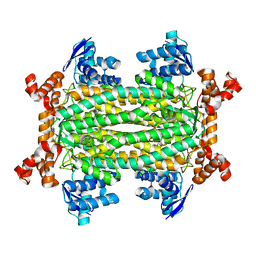
7C1A
 
 | |
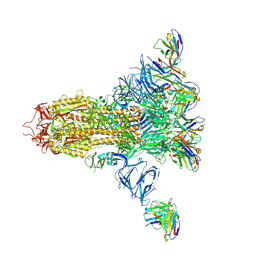
7LXZ
 
 | |
6FYS
 
 | | Structure of single domain antibody SD83 | | Descriptor: | 1,2-ETHANEDIOL, Single domain antibody SD83 | | Authors: | Laursen, N.S, Wilson, I.A. | | Deposit date: | 2018-03-12 | | Release date: | 2018-11-07 | | Last modified: | 2022-03-30 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Universal protection against influenza infection by a multidomain antibody to influenza hemagglutinin.
Science, 362, 2018
|
|
6PS7
 
 | | XFEL A2aR structure by ligand exchange from LUF5843 to ZM241385. | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, (2S)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, 4-{2-[(7-amino-2-furan-2-yl[1,2,4]triazolo[1,5-a][1,3,5]triazin-5-yl)amino]ethyl}phenol, ... | | Authors: | Ishchenko, A, Stauch, B, Han, G.W, Batyuk, A, Shiriaeva, A, Li, C, Zatsepin, N.A, Weierstall, U, Liu, W, Nango, E, Nakane, T, Tanaka, R, Tono, K, Joti, Y, Iwata, S, Moraes, I, Gati, C, Cherezov, C. | | Deposit date: | 2019-07-12 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Toward G protein-coupled receptor structure-based drug design using X-ray lasers.
Iucrj, 6, 2019
|
|
1C8V
 
 | | CRYSTAL STRUCTURE OF THE COMPLEX OF BACTERIAL TRYPTOPHAN SYNTHASE WITH THE TRANSITION STATE ANALOGUE INHIBITOR 4-(2-HYDROXYPHENYLTHIO)-BUTYLPHOSPHONIC ACID | | Descriptor: | 4-(2-HYDROXYPHENYLTHIO)-1-BUTENYLPHOSPHONIC ACID, PYRIDOXAL-5'-PHOSPHATE, SODIUM ION, ... | | Authors: | Sachpatzidis, A, Dealwis, C, Lubetsky, J.B, Liang, P.H, Anderson, K.S, Lolis, E. | | Deposit date: | 1999-07-30 | | Release date: | 2000-01-13 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystallographic studies of phosphonate-based alpha-reaction transition-state analogues complexed to tryptophan synthase.
Biochemistry, 38, 1999
|
|
