1UGJ
 
 | | Solution structure of a murine hypothetical protein from RIKEN cDNA 2310057J16 | | Descriptor: | RIKEN cDNA 2310057J16 protein | | Authors: | Nagashima, T, Hayashi, F, Shirouzu, M, Terada, T, Kigawa, T, Inoue, M, Yabuki, T, Aoki, M, Seki, E, Matsuda, T, Hirota, H, Yoshida, M, Tanaka, A, Osanai, T, Arakawa, T, Carninci, P, Kawai, J, Hayashizaki, Y, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-06-16 | | Release date: | 2004-08-03 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a murine hypothetical protein from RIKEN cDNA 2310057J16
To be Published
|
|
8B47
 
 | | Crystal structure of NAD kinase 1 from Listeria monocytogenes in complex with a cyclic di-adenosine derivative | | Descriptor: | (1~{R},23~{R},24~{S},25~{R})-14-[[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methyl]-7-azanyl-24,25-bis(oxidanyl)-20,20-bis(oxidanylidene)-26-oxa-20$l^{6}-thia-2,4,6,9,14,17,21-heptazatetracyclo[21.2.1.0^{2,10}.0^{3,8}]hexacosa-3,5,7,9-tetraen-11-yn-16-one, NAD kinase 1, PENTAETHYLENE GLYCOL, ... | | Authors: | Gelin, M, Labesse, G. | | Deposit date: | 2022-09-20 | | Release date: | 2023-01-11 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Synthesis and structure-activity relationship studies of original cyclic diadenosine derivatives as nanomolar inhibitors of NAD kinase from pathogenic bacteria.
Eur.J.Med.Chem., 246, 2023
|
|
1U7W
 
 | | Phosphopantothenoylcysteine synthetase from E. coli, CTP-complex | | Descriptor: | CALCIUM ION, CYTIDINE-5'-TRIPHOSPHATE, Coenzyme A biosynthesis bifunctional protein coaBC | | Authors: | Stanitzek, S, Augustin, M.A, Huber, R, Kupke, T, Steinbacher, S. | | Deposit date: | 2004-08-04 | | Release date: | 2004-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Basis of CTP-Dependent Peptide Bond Formation in Coenzyme A Biosynthesis Catalyzed by Escherichia coli PPC Synthetase
STRUCTURE, 12, 2004
|
|
8BBV
 
 | | Coproporphyrin III - LmCpfC complex soaked 2min with Fe2+ | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Coproporphyrin III ferrochelatase, ... | | Authors: | Gabler, T, Hofbauer, S. | | Deposit date: | 2022-10-14 | | Release date: | 2023-01-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Iron insertion into coproporphyrin III-ferrochelatase complex: Evidence for an intermediate distorted catalytic species.
Protein Sci., 32, 2023
|
|
1U8C
 
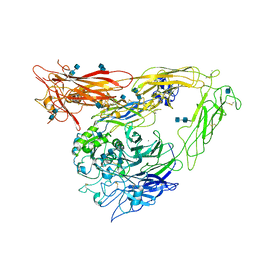 | | A novel adaptation of the integrin PSI domain revealed from its crystal structure | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Xiong, J.P, Stehle, T, Goodman, S.L, Arnaout, M.A. | | Deposit date: | 2004-08-05 | | Release date: | 2004-12-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A novel adaptation of the integrin PSI domain revealed from its crystal structure.
J.Biol.Chem., 279, 2004
|
|
8AVW
 
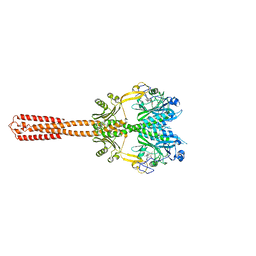 | | Cryo-EM structure of DrBphP in Pr state | | Descriptor: | 3-[2-[(Z)-[3-(2-carboxyethyl)-5-[(Z)-(4-ethenyl-3-methyl-5-oxidanylidene-pyrrol-2-ylidene)methyl]-4-methyl-pyrrol-1-ium -2-ylidene]methyl]-5-[(Z)-[(3E)-3-ethylidene-4-methyl-5-oxidanylidene-pyrrolidin-2-ylidene]methyl]-4-methyl-1H-pyrrol-3- yl]propanoic acid, Bacteriophytochrome,Response regulator | | Authors: | Wahlgren, W.Y, Takala, H, Westenhoff, S. | | Deposit date: | 2022-08-27 | | Release date: | 2022-12-21 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.62 Å) | | Cite: | Structural mechanism of signal transduction in a phytochrome histidine kinase.
Nat Commun, 13, 2022
|
|
1U98
 
 | |
8B3L
 
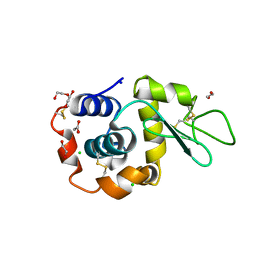 | | Hen Egg White Lysozyme 2s in situ crystallization | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Henkel, A, Galchenkova, M, Yefanov, O, Hakanpaeae, J, Chapman, H.N, Oberthuer, D. | | Deposit date: | 2022-09-16 | | Release date: | 2022-12-21 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | JINXED: just in time crystallization for easy structure determination of biological macromolecules.
Iucrj, 10, 2023
|
|
8BEP
 
 | |
1U9L
 
 | | Structural basis for a NusA- protein N interaction | | Descriptor: | GOLD ION, Lambda N, Transcription elongation protein nusA | | Authors: | Bonin, I, Muehlberger, R, Bourenkov, G.P, Huber, R, Bacher, A, Richter, G, Wahl, M.C. | | Deposit date: | 2004-08-10 | | Release date: | 2004-08-31 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for the interaction of Escherichia coli NusA with protein N of phage lambda
Proc.Natl.Acad.Sci.Usa, 101, 2004
|
|
8BI0
 
 | | ABCG2 turnover-2 state with tariquidar bound | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Broad substrate specificity ATP-binding cassette transporter ABCG2, CHOLESTEROL, ... | | Authors: | Yu, Q, Kowal, J, Tajkhorshid, E, Locher, K.P. | | Deposit date: | 2022-11-01 | | Release date: | 2023-01-11 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Differential dynamics and direct interaction of bound ligands with lipids in multidrug transporter ABCG2.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
1U9T
 
 | |
1UAA
 
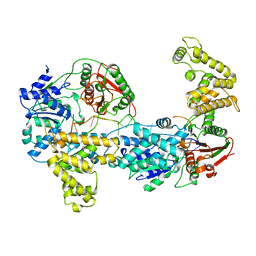 | | E. COLI REP HELICASE/DNA COMPLEX | | Descriptor: | DNA (5'-D(*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*TP*T)-3'), PROTEIN (ATP-DEPENDENT DNA HELICASE REP.) | | Authors: | Korolev, S, Waksman, G. | | Deposit date: | 1997-06-30 | | Release date: | 1998-07-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Major domain swiveling revealed by the crystal structures of complexes of E. coli Rep helicase bound to single-stranded DNA and ADP.
Cell(Cambridge,Mass.), 90, 1997
|
|
8B5C
 
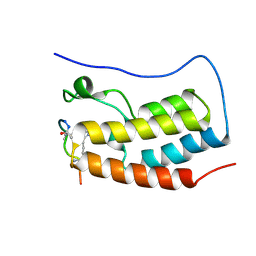 | | Human BRD4 bromdomain 1 in complex with a H4 peptide containing ApmTri (H4K5/8ApmTri) | | Descriptor: | Bromodomain-containing protein 4, H4K5/8ApmTri | | Authors: | Braun, M.B, Bartlick, N, Stehle, T. | | Deposit date: | 2022-09-22 | | Release date: | 2023-01-11 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Synthesis, Biochemical Characterization, and Genetic Encoding of a 1,2,4-Triazole Amino Acid as an Acetyllysine Mimic for Bromodomains of the BET Family.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
1UB6
 
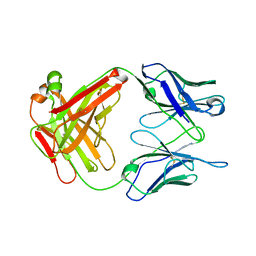 | | Crystal structure of Antibody 19G2 with sera ligand | | Descriptor: | antibody 19G2, alpha chain, beta chain | | Authors: | Beuscher, A.B, Wirsching, P, Lerner, R.A, Janda, K, Stevens, R.C. | | Deposit date: | 2003-03-30 | | Release date: | 2004-04-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure and Dynamics of Blue Fluorescent Antibody 19G2 at Blue and Violet Fluorescent Temperatures
To be published
|
|
8BC7
 
 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex an aminoglutarimide degron peptide | | Descriptor: | Cereblon isoform 4, PHE-PHE-GLU-GLN-MET-GLN-QCI, S-Thalidomide, ... | | Authors: | Heim, C, Albrecht, R, Spring, A.K, Hartmann, M.D. | | Deposit date: | 2022-10-15 | | Release date: | 2023-01-11 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.719 Å) | | Cite: | Identification and structural basis of C-terminal cyclic imides as natural degrons for cereblon.
Biochem.Biophys.Res.Commun., 637, 2022
|
|
1UBR
 
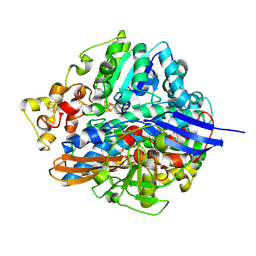 | | Three-dimensional Structure of The Carbon Monoxide Complex of [NiFe]hydrogenase From Desulufovibrio vulgaris Miyazaki F | | Descriptor: | (MU-SULPHIDO)-BIS(MU-CYS,S)-[TRICARBONYLIRON-DI-(CYS,S)NICKEL(II)](FE-NI), CARBON MONOXIDE, FE3-S4 CLUSTER, ... | | Authors: | Ogata, H, Mizoguchi, Y, Mizuno, N, Miki, K, Adachi, S, Yasuoka, N, Yagi, T, Yamauchi, O, Hirota, S, Higuchi, Y. | | Deposit date: | 2003-04-04 | | Release date: | 2003-04-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Structural Studies of the Carbon Monoxide Complex of [NiFe]hydrogenase from Desulfovibrio vulgaris Miyazaki F: Suggestion for the Initial Activation Site for Dihydrogen
J.Am.Chem.Soc., 124, 2002
|
|
8BGP
 
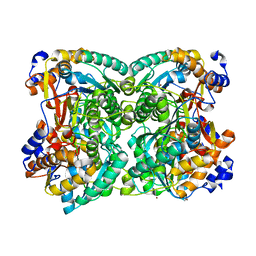 | | N,N-diacetylchitobiose deacetylase from Pyrococcus chitonophagus anomalous data | | Descriptor: | Diacetylchitobiose deacetylase, ZINC ION | | Authors: | Rypniewski, W, Biniek-Antosiak, K, Bejger, M. | | Deposit date: | 2022-10-28 | | Release date: | 2023-01-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structural, Thermodynamic and Enzymatic Characterization of N , N -Diacetylchitobiose Deacetylase from Pyrococcus chitonophagus.
Int J Mol Sci, 23, 2022
|
|
1UCG
 
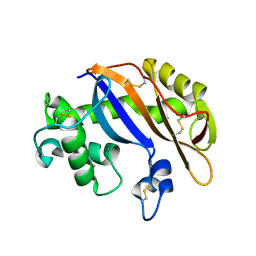 | | Crystal structure of Ribonuclease MC1 N71T mutant | | Descriptor: | MANGANESE (II) ION, Ribonuclease MC | | Authors: | Suzuki, A, Numata, T, Yao, M, Tanaka, I, Kimura, M. | | Deposit date: | 2003-04-14 | | Release date: | 2003-04-29 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal structures of the ribonuclease MC1 mutants N71T and N71S in complex with 5'-GMP: structural basis for alterations in substrate specificity
Biochemistry, 42, 2003
|
|
8BEE
 
 | |
8BBE
 
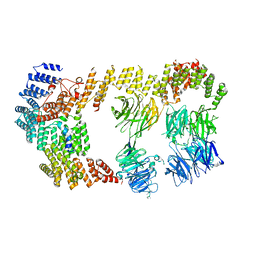 | | Structure of the IFT-A complex; IFT-A2 module | | Descriptor: | Intraflagellar transport protein 122 homolog, Intraflagellar transport protein 43 homolog, SNAP-tag,Tetratricopeptide repeat protein 21B, ... | | Authors: | Hesketh, S.J, Mukhopadhyay, A.G, Nakamura, D, Toropova, K, Roberts, A.J. | | Deposit date: | 2022-10-12 | | Release date: | 2022-12-21 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | IFT-A structure reveals carriages for membrane protein transport into cilia.
Cell, 185, 2022
|
|
1UCT
 
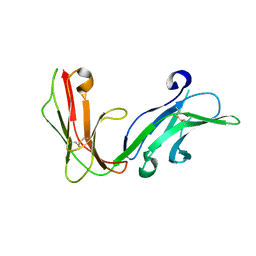 | | Crystal structure of the extracellular fragment of Fc alpha Receptor I (CD89) | | Descriptor: | Immunoglobulin alpha Fc receptor | | Authors: | Ding, Y, Xu, G, Yang, M, Zhang, W, Rao, Z. | | Deposit date: | 2003-04-21 | | Release date: | 2003-07-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of the Ectodomain of Human Fc{alpha}RI.
J.Biol.Chem., 278, 2003
|
|
8B1T
 
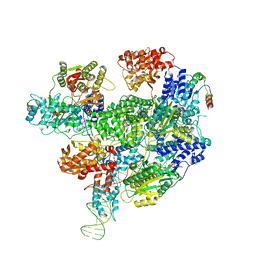 | | RecBCD-DNA in complex with the phage protein Abc2 | | Descriptor: | Anti-RecBCD protein 2, DNA (70-MER), MAGNESIUM ION, ... | | Authors: | Wilkinson, M, Wilkinson, O.J, Feyerherm, C, Fletcher, E.E, Wigley, D.B, Dillingham, M.S. | | Deposit date: | 2022-09-12 | | Release date: | 2022-12-28 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structures of RecBCD in complex with phage-encoded inhibitor proteins reveal distinctive strategies for evasion of a bacterial immunity hub.
Elife, 11, 2022
|
|
1UD5
 
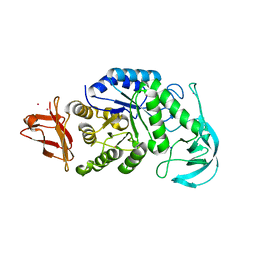 | | Crystal structure of AmyK38 with rubidium ion | | Descriptor: | RUBIDIUM ION, SODIUM ION, amylase | | Authors: | Nonaka, T, Fujihashi, M, Kita, A, Hagihara, H, Ozaki, K, Ito, S, Miki, K. | | Deposit date: | 2003-04-28 | | Release date: | 2003-07-22 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of calcium-free alpha-amylase from Bacillus sp. strain KSM-K38 (AmyK38) and its sodium ion binding sites
J.Biol.Chem., 278, 2003
|
|
1UE6
 
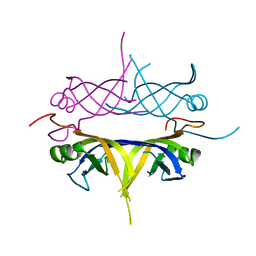 | | Crystal structure of the single-stranded dna-binding protein from mycobacterium tuberculosis | | Descriptor: | Single-strand binding protein | | Authors: | Saikrishnan, K, Jeyakanthan, J, Venkatesh, J, Acharya, N, Sekar, K, Varshney, U, Vijayan, M, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2003-05-09 | | Release date: | 2004-02-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure of Mycobacterium tuberculosis single-stranded DNA-binding protein. Variability in quaternary structure and its implications
J.MOL.BIOL., 331, 2003
|
|
