5CVF
 
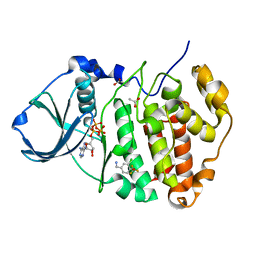 | | Crystal Structure of CK2alpha with Compound 5 bound | | Descriptor: | 1-[3-chloro-4-(trifluoromethoxy)phenyl]methanamine, ACETATE ION, ADENOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Brear, P, De Fusco, C, Georgiou, K.H, Spring, D, Hyvonen, M. | | Deposit date: | 2015-07-26 | | Release date: | 2016-07-27 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Specific inhibition of CK2 alpha from an anchor outside the active site.
Chem Sci, 7, 2016
|
|
8W0J
 
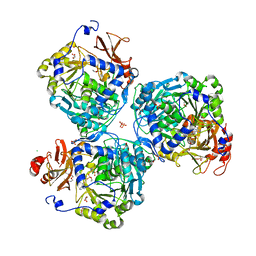 | |
8S6M
 
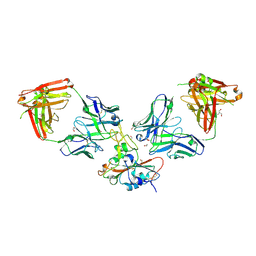 | | SARS-CoV-2 BQ.1.1 RBD bound to the S2V29 and the S2H97 Fab fragments | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Errico, J.M, Park, Y.J, Rietz, T, Czudnochowski, N, Nix, J.C, Cameroni, E, Corti, D, Snell, G, Marco, A.D, Pinto, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Veesler, D. | | Deposit date: | 2024-02-28 | | Release date: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | A potent pan-sarbecovirus neutralizing antibody resilient to epitope diversification.
Cell, 2024
|
|
8W0H
 
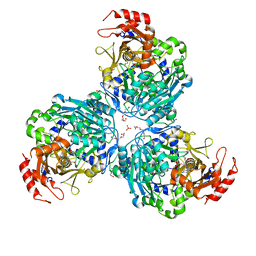 | |
7PT5
 
 | |
5DIO
 
 | |
8OVV
 
 | |
5DIV
 
 | | The Fk1 domain of FKBP51 in complex with the new synthetic ligand (S)-N-(1-carbamoylcyclopentyl)-1-((S)-2-cyclohexyl-2-(3,4,5-trimethoxyphenyl)acetyl)piperidine-2-carboxamide | | Descriptor: | (2S)-N-(1-carbamoylcyclopentyl)-1-[(2S)-2-cyclohexyl-2-(3,4,5-trimethoxyphenyl)acetyl]piperidine-2-carboxamide, Peptidyl-prolyl cis-trans isomerase FKBP5 | | Authors: | Gaali, S, Feng, X, Sippel, C, Bracher, A, Hausch, F. | | Deposit date: | 2015-09-01 | | Release date: | 2016-03-23 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Rapid, Structure-Based Exploration of Pipecolic Acid Amides as Novel Selective Antagonists of the FK506-Binding Protein 51.
J.Med.Chem., 59, 2016
|
|
7PW1
 
 | | Crystal structure of ancestral haloalkane dehalogenase AncLinB-DmbA | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Haloalkane dehalogenase, ... | | Authors: | Mazur, A, Grinkevich, P, Prudnikova, T. | | Deposit date: | 2021-10-05 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural Analysis of the Ancestral Haloalkane Dehalogenase AncLinB-DmbA.
Int J Mol Sci, 22, 2021
|
|
5UJ8
 
 | | Human Origin Recognition Complex subunits 2 and 3 | | Descriptor: | Origin recognition complex subunit 2, Origin recognition complex subunit 3 | | Authors: | Tocilj, A, On, K.F, Elkayam, E, Joshua-Tor, L. | | Deposit date: | 2017-01-17 | | Release date: | 2017-02-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | Structure of the active form of human Origin Recognition Complex and its ATPase motor module.
Elife, 6, 2017
|
|
7VPY
 
 | | Crystal structure of the neutralizing nanobody P86 against SARS-CoV-2 | | Descriptor: | 1,2-ETHANEDIOL, Nanobody, SULFATE ION | | Authors: | Maeda, R, Fujita, J, Konishi, Y, Kazuma, Y, Yamazaki, H, Anzai, I, Yamaguchi, K, Kasai, K, Nagata, K, Yamaoka, Y, Miyakawa, K, Ryo, A, Shirakawa, K, Makino, F, Matsuura, Y, Inoue, T, Imura, A, Namba, K, Takaori-Kondo, A. | | Deposit date: | 2021-10-18 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | A panel of nanobodies recognizing conserved hidden clefts of all SARS-CoV-2 spike variants including Omicron.
Commun Biol, 5, 2022
|
|
8OM6
 
 | |
5UK8
 
 | | The co-structure of (R)-4-(6-(1-(cyclopropylsulfonyl)cyclopropyl)-2-(1H-indol-4-yl)pyrimidin-4-yl)-3-methylmorpholine and a rationally designed PI3K-alpha mutant that mimics ATR | | Descriptor: | (R)-4-(6-(1-(cyclopropylsulfonyl)cyclopropyl)-2-(1H-indol-4-yl)pyrimidin-4-yl)-3-methylmorpholine, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Knapp, M.S, Mamo, M, Elling, R.A. | | Deposit date: | 2017-01-20 | | Release date: | 2017-06-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Rationally Designed PI3K alpha Mutants to Mimic ATR and Their Use to Understand Binding Specificity of ATR Inhibitors.
J. Mol. Biol., 429, 2017
|
|
8P61
 
 | | Crystal structure of O'nyong'nyong virus capsid protease (106-256) | | Descriptor: | Capsid protein, GLYCEROL, SULFATE ION | | Authors: | Plewka, J, Chykunova, Y, Wilk, P, Sienczyk, M, Dubin, G, Pyrc, K. | | Deposit date: | 2023-05-24 | | Release date: | 2024-03-06 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Autoinhibition of suicidal capsid protease from O'nyong'nyong virus.
Int.J.Biol.Macromol., 262, 2024
|
|
8W0M
 
 | |
5HWF
 
 | |
8WD5
 
 | |
8STL
 
 | | Crystal Structure of Nanobody PIK3_Nb16 against wild-type PI3Kalpha | | Descriptor: | Nanobody PIK3_Nb16, SULFATE ION | | Authors: | Nwafor, J.N, Srinivasan, L, Chen, Z, Gabelli, S.B, Iheanacho, A, Alzogaray, V, Klinke, S. | | Deposit date: | 2023-05-10 | | Release date: | 2024-05-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Development of isoform specific nanobodies for Class I PI3Ks
To be published
|
|
8STB
 
 | | The structure of abxF, an enzyme catalyzing the formation of the chiral spiroketal of an anthrabenzoxocinone antibiotic, (-)-ABX | | Descriptor: | CHLORIDE ION, GLYCEROL, Glyoxalase, ... | | Authors: | Luo, Z, Jia, X, Yan, X, Qu, X, Kobe, B. | | Deposit date: | 2023-05-09 | | Release date: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | The crystal structure of abxF, an enzyme catalyzing the formation of the chiral spiroketal of an anthrabenzoxocinone antibiotic, (-)-ABX.
To Be Published
|
|
8OZ4
 
 | | Populus tremula stable protein 1 with an alternate crystal lattice | | Descriptor: | Stable protein 1 | | Authors: | Sklyar, J, Zeibaq, Y, Bachar, O, Yehezkeli, O, Adir, N. | | Deposit date: | 2023-05-08 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | A Bioengineered Stable Protein 1-Hemin Complex with Enhanced Peroxidase-Like Catalytic Properties
Small Science, 2024
|
|
7VHF
 
 | | Crystal structure of the STX2a complexed with RRA peptide | | Descriptor: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, GLYCEROL, RRA peptide, ... | | Authors: | Senda, M, Takahashi, M, Nishikawa, K, Senda, T. | | Deposit date: | 2021-09-22 | | Release date: | 2022-07-20 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | A unique peptide-based pharmacophore identifies an inhibitory compound against the A-subunit of Shiga toxin.
Sci Rep, 12, 2022
|
|
8P37
 
 | | Structure a catalytically inactive mutant of the IMP dehydrogenase related protein GUAB3 from Synechocystis PCC 6803 | | Descriptor: | IMP dehydrogenase subunit, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, XANTHOSINE-5'-MONOPHOSPHATE | | Authors: | Hernandez-Gomez, A, Fernandez-Justel, D, Buey, R.M. | | Deposit date: | 2023-05-17 | | Release date: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.219 Å) | | Cite: | GuaB3, an overlooked enzyme in cyanobacteria's toolbox that sheds light on IMP dehydrogenase evolution.
Structure, 31, 2023
|
|
5ULF
 
 | |
7YM0
 
 | | Lysoplasmalogen-specific phospholipase D (LyPls-PLD) with Ca2+ | | Descriptor: | CALCIUM ION, Lysoplasmalogenase | | Authors: | Yasutake, Y, Sakasegawa, S, Sugimori, D, Murayama, K. | | Deposit date: | 2022-07-27 | | Release date: | 2023-01-04 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Structural basis for the substrate specificity switching of lysoplasmalogen-specific phospholipase D from Thermocrispum sp. RD004668.
Biosci.Biotechnol.Biochem., 87, 2022
|
|
8W0L
 
 | |
