4CJ1
 
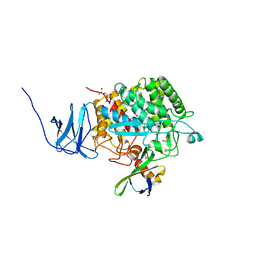 | | Crystal structure of CelD in complex with affitin H3 | | Descriptor: | CALCIUM ION, ENDOGLUCANASE D, GLYCEROL, ... | | Authors: | Correa, A, Pacheco, S, Mechaly, A.E, Obal, G, Behar, G, Mouratou, B, Oppezzo, P, Alzari, P.M, Pecorari, F. | | Deposit date: | 2013-12-18 | | Release date: | 2014-05-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Potent and Specific Inhibition of Glycosidases by Small Artificial Binding Proteins (Affitins)
Plos One, 9, 2014
|
|
2N18
 
 | |
5TOT
 
 | |
5U74
 
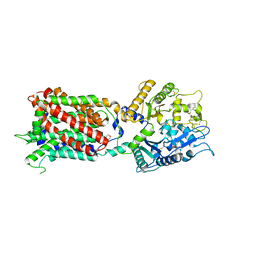 | | Structure of human Niemann-Pick C1 protein | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Li, X. | | Deposit date: | 2016-12-11 | | Release date: | 2017-10-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.335 Å) | | Cite: | 3.3 angstrom structure of Niemann-Pick C1 protein reveals insights into the function of the C-terminal luminal domain in cholesterol transport.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
8EL9
 
 | | Cryo-EM structure of human catalase | | Descriptor: | Catalase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Su, C.C. | | Deposit date: | 2022-09-23 | | Release date: | 2023-10-04 | | Method: | ELECTRON MICROSCOPY (2.27 Å) | | Cite: | Cryo-EM structure of human catalase
To Be Published
|
|
6V3A
 
 | |
2N0C
 
 | | NMR structure of Neuromedin C in 10% TFE | | Descriptor: | Neuromedin C (NMC) | | Authors: | Adrover, M, Sanchis, P, Vilanova, B, Pauwels, K, Martorell, G, Perez, J. | | Deposit date: | 2015-03-05 | | Release date: | 2015-10-21 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Conformational ensembles of neuromedin C reveal a progressive coil-helix transition within a binding-induced folding mechanism.
RSC ADV, 5, 2015
|
|
6V62
 
 | | SETD3 double mutant (N255F/W273A) in Complex with an Actin Peptide with His73 Replaced with Lysine | | Descriptor: | 1,2-ETHANEDIOL, Actin, cytoplasmic 1, ... | | Authors: | Dai, S, Horton, J.R, Cheng, X. | | Deposit date: | 2019-12-04 | | Release date: | 2020-01-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.36 Å) | | Cite: | An engineered variant of SETD3 methyltransferase alters target specificity from histidine to lysine methylation.
J.Biol.Chem., 295, 2020
|
|
5UA5
 
 | |
5UCU
 
 | | STRUCTURAL AND MECHANISTIC INSIGHTS INTO HEMOGLOBIN-CATALYZED HYDROGEN SULFIDE OXIDATION AND THE FATE OF POLYSULFIDE PRODUCTS | | Descriptor: | HYDROSULFURIC ACID, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Vitvitsky, V, Yadav, P.K, An, S, Seravalli, J, Cho, U.-S, Banerjee, R. | | Deposit date: | 2016-12-22 | | Release date: | 2017-03-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | Structural and Mechanistic Insights into Hemoglobin-catalyzed Hydrogen Sulfide Oxidation and the Fate of Polysulfide Products.
J. Biol. Chem., 292, 2017
|
|
8EL0
 
 | | Structure of MBP-Mcl-1 in complex with a macrocyclic compound | | Descriptor: | (7R,20P)-18-chloro-1-(4-fluorophenyl)-10-{[(2M)-2-(2-methoxyphenyl)pyrimidin-4-yl]methoxy}-19-methyl-15-[2-(4-methylpiperazin-1-yl)ethyl]-7,8,15,16-tetrahydro-14H-17,20-etheno-9,13-(metheno)-6-oxa-2-thia-3,5,15-triazacyclooctadeca[1,2,3-cd]indene-7-carboxylic acid, Maltose/maltodextrin-binding periplasmic protein,Induced myeloid leukemia cell differentiation protein Mcl-1, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose | | Authors: | Judge, R.A, Judd, A.S, Souers, A.J. | | Deposit date: | 2022-09-22 | | Release date: | 2023-10-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.917 Å) | | Cite: | Selective MCL-1 inhibitor ABBV-467 is efficacious in tumor models but is associated with cardiac troponin increases in patients.
Commun Med (Lond), 3, 2023
|
|
2N0B
 
 | | NMR structure of Neuromedin C in aqueous solution | | Descriptor: | Neuromedin C (NMC) | | Authors: | Adrover, M, Sanchis, P, Vilanova, B, Pauwels, K, Martorell, G, Perez, J. | | Deposit date: | 2015-03-05 | | Release date: | 2015-10-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Conformational ensembles of neuromedin C reveal a progressive coil-helix transition within a binding-induced folding mechanism.
RSC ADV, 5, 2015
|
|
6V8K
 
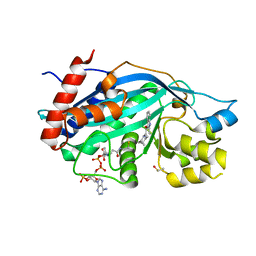 | |
4CI3
 
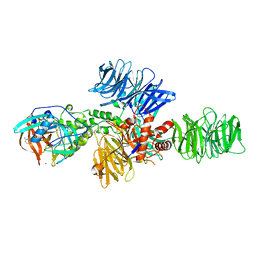 | | Structure of the DDB1-CRBN E3 ubiquitin ligase bound to Pomalidomide | | Descriptor: | DNA DAMAGE-BINDING PROTEIN 1, PROTEIN CEREBLON, S-Pomalidomide, ... | | Authors: | Fischer, E.S, Boehm, K, Thoma, N.H. | | Deposit date: | 2013-12-05 | | Release date: | 2014-07-16 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Structure of the Ddb1-Crbn E3 Ubiquitin Ligase in Complex with Thalidomide.
Nature, 512, 2014
|
|
6SJF
 
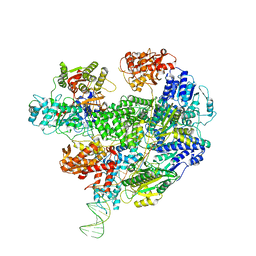 | | Cryo-EM structure of the RecBCD Chi unrecognised complex | | Descriptor: | Forked DNA substrate, RecBCD enzyme subunit RecB, RecBCD enzyme subunit RecC, ... | | Authors: | Cheng, K, Wilkinson, M, Wigley, D.B. | | Deposit date: | 2019-08-13 | | Release date: | 2020-01-01 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | A conformational switch in response to Chi converts RecBCD from phage destruction to DNA repair.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6S8G
 
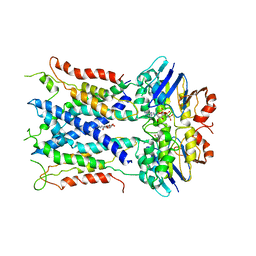 | | Cryo-EM structure of LptB2FGC in complex with AMP-PNP | | Descriptor: | Inner membrane protein yjgQ, LPS export ABC transporter permease LptF, Lipopolysaccharide ABC transporter, ... | | Authors: | Tang, X.D, Chang, S.H, Luo, Q.H, Zhang, Z.Y, Qiao, W, Xu, C.H, Zhang, C.B, Niu, Y, Yang, W.X, Wang, T, Zhang, Z.B, Zhu, X.F, Dong, C.J, Zhang, X, Dong, H.H. | | Deposit date: | 2019-07-10 | | Release date: | 2019-09-25 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structures of lipopolysaccharide transporter LptB2FGC in lipopolysaccharide or AMP-PNP-bound states reveal its transport mechanism.
Nat Commun, 10, 2019
|
|
4CXR
 
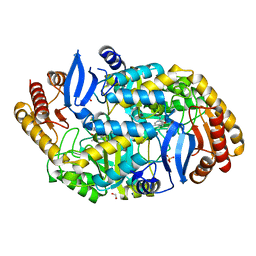 | | Mycobaterium tuberculosis transaminase BioA complexed with 1-(1,3- benzothiazol-2-yl)methanamine | | Descriptor: | 1,2-ETHANEDIOL, 1-(1,3-benzothiazol-2-yl)methanamine, ADENOSYLMETHIONINE-8-AMINO-7-OXONONANOATE AMINOTRANSFERASE, ... | | Authors: | Dai, R, Geders, T.W, Finzel, B.C. | | Deposit date: | 2014-04-08 | | Release date: | 2014-04-23 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Inhibition of Mycobacterium Tuberculosis Transaminase Bioa by Aryl Hydrazines and Hydrazides.
Chembiochem, 15, 2014
|
|
2MWF
 
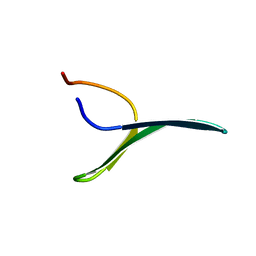 | |
5UHN
 
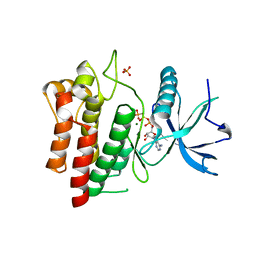 | |
6SPF
 
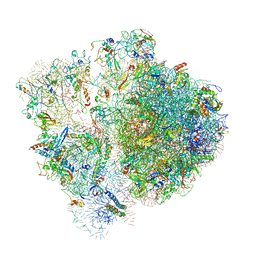 | | Pseudomonas aeruginosa 70s ribosome from an aminoglycoside resistant clinical isolate | | Descriptor: | 16S rRNA, 30S ribosomal protein S10, 30S ribosomal protein S11, ... | | Authors: | Halfon, Y, Jimenez-Fernande, A, La Ros, R, Espinos, R, Krogh Johansen, H, Matzov, D, Eyal, Z, Bashan, A, Zimmerman, E, Belousoff, M, Molin, S, Yonath, A. | | Deposit date: | 2019-09-01 | | Release date: | 2019-10-23 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structure ofPseudomonas aeruginosaribosomes from an aminoglycoside-resistant clinical isolate.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4COB
 
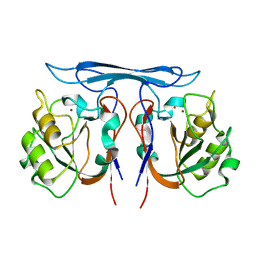 | | Crystal structure kynurenine formamidase from Pseudomonas aeruginosa | | Descriptor: | KYNURENINE FORMAMIDASE, ZINC ION | | Authors: | Diaz-Saez, L, Srikannathasan, V, Zoltner, M, Hunter, W.N. | | Deposit date: | 2014-01-28 | | Release date: | 2014-04-02 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structure of Bacterial Kynurenine Formamidase Reveals a Crowded Binuclear-Zinc Catalytic Site Primed to Generate a Potent Nucleophile.
Biochem.J., 462, 2014
|
|
6S8V
 
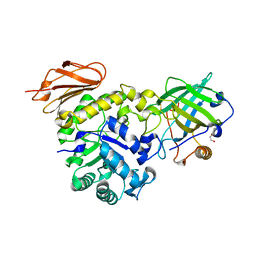 | | Structure of the high affinity Anticalin P3D11 in complex with the human CD98 heavy chain ectodomain | | Descriptor: | 1,2-ETHANEDIOL, 4F2 cell-surface antigen heavy chain, Neutrophil gelatinase-associated lipocalin | | Authors: | Schiefner, A, Deuschle, F.-C, Skerra, A. | | Deposit date: | 2019-07-10 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Development of a high affinity Anticalin®directed against human CD98hc for theranostic applications.
Theranostics, 10, 2020
|
|
5UA4
 
 | |
6V01
 
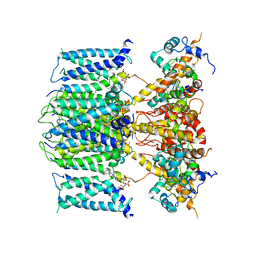 | | structure of human KCNQ1-KCNE3-CaM complex with PIP2 | | Descriptor: | CALCIUM ION, Calmodulin-1, Potassium voltage-gated channel subfamily E member 3, ... | | Authors: | Mackinnon, R, Sun, J. | | Deposit date: | 2019-11-18 | | Release date: | 2019-12-04 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Basis of Human KCNQ1 Modulation and Gating.
Cell, 180, 2020
|
|
6VHQ
 
 | | Crystal structure of Bacillus subtilis levansucrase (D86A/E342A) in complex with oligosaccharides | | Descriptor: | BROMIDE ION, CALCIUM ION, Glycoside hydrolase family 68 protein, ... | | Authors: | Diaz-Vilchis, A, Raga-Carbajal, E, Rojas-Trejo, S, Olvera, C, Rudino-Pinera, E. | | Deposit date: | 2020-01-10 | | Release date: | 2021-01-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.047 Å) | | Cite: | The molecular basis of the nonprocessive elongation mechanism in levansucrases.
J.Biol.Chem., 296, 2020
|
|
