4TW3
 
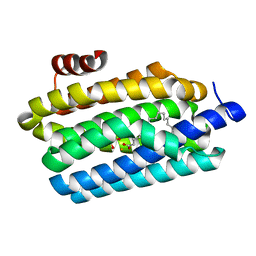 | | Insights into Substrate and Metal Binding from the Crystal Structure of Cyanobacterial Aldehyde Deformylating Oxygenase with Substrate Bound | | Descriptor: | 1,2-ETHANEDIOL, Aldehyde decarbonylase, FE (III) ION, ... | | Authors: | Buer, B.C, Paul, B, Das, D, Stuckey, J.A, Marsh, E.N.G. | | Deposit date: | 2014-06-29 | | Release date: | 2014-10-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Insights into substrate and metal binding from the crystal structure of cyanobacterial aldehyde deformylating oxygenase with substrate bound.
Acs Chem.Biol., 9, 2014
|
|
7DAB
 
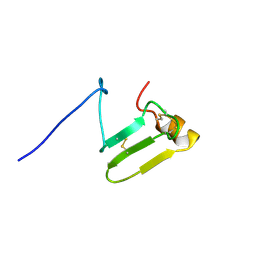 | | plant peptide hormone | | Descriptor: | Maternally expressed gene14 | | Authors: | Umetsu, Y, Ohki, S. | | Deposit date: | 2020-10-16 | | Release date: | 2021-10-20 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | plant peptide hormone
To Be Published
|
|
7DD4
 
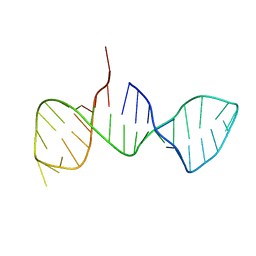 | |
4TQG
 
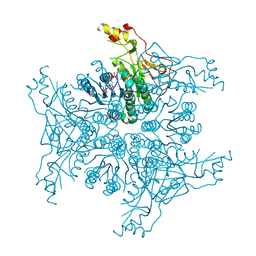 | | Crystal structure of Megavirus UDP-GlcNAc 4,6-dehydratase, 5-epimerase Mg534 | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Putative dTDP-d-glucose 4 6-dehydratase | | Authors: | Jeudy, S, Piacente, F, De Castro, C, Molinaro, A, Salis, A, Damonte, G, Bernardi, C, Tonetti, M, Claverie, J.M, Abergel, C. | | Deposit date: | 2014-06-11 | | Release date: | 2014-06-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Giant Virus Megavirus chilensis Encodes the Biosynthetic Pathway for Uncommon Acetamido Sugars.
J.Biol.Chem., 289, 2014
|
|
7DTK
 
 | |
7DTJ
 
 | |
4TZ8
 
 | | Structure of human ATAD2 bromodomain bound to fragment inhibitor | | Descriptor: | 2-amino-7,7-dimethyl-5,6,7,8-tetrahydro-4H-[1,3]thiazolo[5,4-c]azepin-4-one, ATPase family AAA domain-containing protein 2, CHLORIDE ION, ... | | Authors: | Harner, M.J, Chauder, B.A, Phan, J, Fesik, S.W. | | Deposit date: | 2014-07-09 | | Release date: | 2014-10-29 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Fragment-Based Screening of the Bromodomain of ATAD2.
J.Med.Chem., 57, 2014
|
|
7E0E
 
 | |
5OFQ
 
 | |
6G6J
 
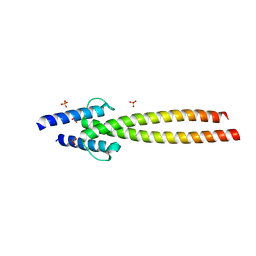 | | The crystal structures of Human MYC:MAX bHLHZip complex | | Descriptor: | Myc proto-oncogene protein, Protein max, SULFATE ION | | Authors: | Allen, M.D, Zinzalla, G. | | Deposit date: | 2018-04-01 | | Release date: | 2019-04-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal Structures and Nuclear Magnetic Resonance Studies of the Apo Form of the c-MYC:MAX bHLHZip Complex Reveal a Helical Basic Region in the Absence of DNA.
Biochemistry, 58, 2019
|
|
4RMW
 
 | |
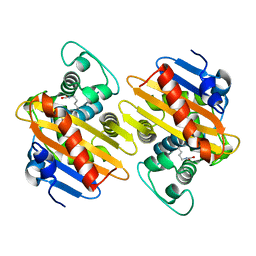
5OFT
 
 | |
6G6K
 
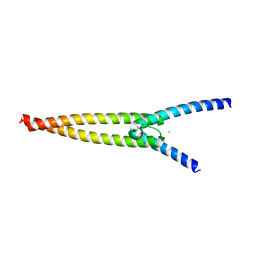 | | The crystal structures of Human MYC:MAX bHLHZip complex | | Descriptor: | CHLORIDE ION, Myc proto-oncogene protein, Protein max | | Authors: | Allen, M.D, Zinzalla, G. | | Deposit date: | 2018-04-01 | | Release date: | 2019-04-10 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal Structures and Nuclear Magnetic Resonance Studies of the Apo Form of the c-MYC:MAX bHLHZip Complex Reveal a Helical Basic Region in the Absence of DNA.
Biochemistry, 58, 2019
|
|
4TYT
 
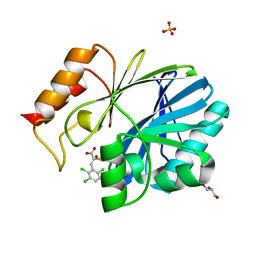 | | Crystal Structure of BcII metallo-beta-lactamase in complex with ML302F | | Descriptor: | (2Z)-2-sulfanyl-3-(2,3,6-trichlorophenyl)prop-2-enoic acid, Beta-lactamase 2, GLYCEROL, ... | | Authors: | Brem, J, van Berkel, S.S, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2014-07-09 | | Release date: | 2014-11-26 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.799 Å) | | Cite: | Rhodanine hydrolysis leads to potent thioenolate mediated metallo-beta-lactamase inhibition.
Nat.Chem., 6, 2014
|
|
4TZ2
 
 | | Fragment-Based Screening of the Bromodomain of ATAD2 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 3-(5-phenyl-4H-1,2,4-triazol-3-yl)aniline, ATPase family AAA domain-containing protein 2, ... | | Authors: | Harner, M.J, Chauder, B.A, Phan, J, Fesik, S.W. | | Deposit date: | 2014-07-09 | | Release date: | 2014-10-29 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Fragment-Based Screening of the Bromodomain of ATAD2.
J.Med.Chem., 57, 2014
|
|
4TYL
 
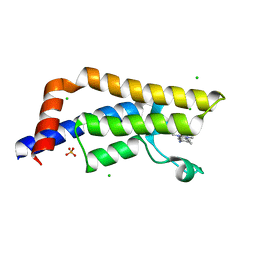 | | Fragment-Based Screening of the Bromodomain of ATAD2 | | Descriptor: | 5-amino-1,3,6-trimethyl-1,3-dihydro-2H-benzimidazol-2-one, ATPase family AAA domain-containing protein 2, CHLORIDE ION, ... | | Authors: | Harner, M.J, Chauder, B.A, Phan, J, Fesik, S.W. | | Deposit date: | 2014-07-08 | | Release date: | 2014-10-29 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Fragment-Based Screening of the Bromodomain of ATAD2.
J.Med.Chem., 57, 2014
|
|
4UCM
 
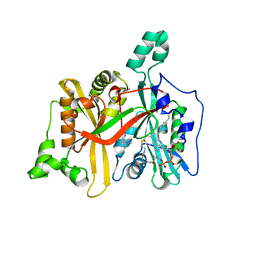 | |
4U9I
 
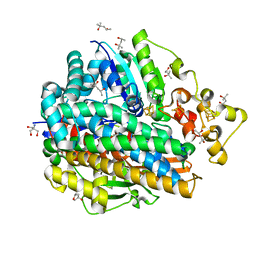 | | High Resolution Structure Of The Ni-R State Of [Nife]Hydrogenase From Desulufovibrio Vulgaris Miyazaki F | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FE3-S4 CLUSTER, ... | | Authors: | Ogata, H, Nishikawa, K, Lubitz, W. | | Deposit date: | 2014-08-06 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.06 Å) | | Cite: | Hydrogens detected by subatomic resolution protein crystallography in a [NiFe] hydrogenase.
Nature, 520, 2015
|
|
5OQZ
 
 | |
4U5M
 
 | | Structure of a left-handed DNA G-quadruplex | | Descriptor: | DNA (28-MER), MAGNESIUM ION, POTASSIUM ION | | Authors: | Schmitt, E, Mechulam, Y, Phan, A.T, Brahim, H, Chung, W.J, Lim, K.W. | | Deposit date: | 2014-07-25 | | Release date: | 2015-02-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of a left-handed DNA G-quadruplex.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
2WF6
 
 | | Structure of Beta-Phosphoglucomutase inhibited with Glucose-6-phosphate and Aluminium tetrafluoride | | Descriptor: | 6-O-phosphono-beta-D-glucopyranose, BETA-PHOSPHOGLUCOMUTASE, MAGNESIUM ION, ... | | Authors: | Bowler, M.W, Baxter, N.J, Webster, C.E, Pollard, S, Alizadeh, T, Hounslow, A.M, Cliff, M.J, Bermel, W, Williams, N.H, Hollfelder, F, Blackburn, G.M, Waltho, J.P. | | Deposit date: | 2009-04-03 | | Release date: | 2010-05-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Atomic details of near-transition state conformers for enzyme phosphoryl transfer revealed by MgF-3 rather than by phosphoranes.
Proc. Natl. Acad. Sci. U.S.A., 107, 2010
|
|
2WF5
 
 | | Structure of Beta-Phosphoglucomutase inhibited with Glucose-6-phosphate and trifluoromagnesate | | Descriptor: | 6-O-phosphono-beta-D-glucopyranose, BETA-PHOSPHOGLUCOMUTASE, MAGNESIUM ION, ... | | Authors: | Bowler, M.W, Baxter, N.J, Webster, C.E, Hounslow, A.M, Cliff, M.J, Williams, N.H, Hollfelder, F, Gamblin, S, Blackburn, G.M, Waltho, J.P. | | Deposit date: | 2009-04-03 | | Release date: | 2009-09-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Atomic details of near-transition state conformers for enzyme phosphoryl transfer revealed by MgF-3 rather than by phosphoranes.
Proc. Natl. Acad. Sci. U.S.A., 107, 2010
|
|
5OOQ
 
 | | Structure of the Mtr4 Nop53 Complex | | Descriptor: | ATP-dependent RNA helicase DOB1, Ribosome biogenesis protein NOP53, SULFATE ION | | Authors: | Falk, S, Basquin, J, Conti, E. | | Deposit date: | 2017-08-08 | | Release date: | 2017-09-20 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural insights into the interaction of the nuclear exosome helicase Mtr4 with the preribosomal protein Nop53.
RNA, 23, 2017
|
|
4UP5
 
 | | Crystal structure of the Pygo2 PHD finger in complex with the B9L HD1 domain and a chemical fragment | | Descriptor: | 6-methoxy-1,3-benzothiazol-2-amine, PYGOPUS HOMOLOG 2, B-CELL CLL/LYMPHOMA 9-LIKE PROTEIN, ... | | Authors: | Miller, T.C.R, Fiedler, M, Rutherford, T.J, Birchall, K, Chugh, J, Bienz, M. | | Deposit date: | 2014-06-12 | | Release date: | 2014-11-05 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Competitive Binding of a Benzimidazole to the Histone-Binding Pocket of the Pygo Phd Finger.
Acs Chem.Biol., 9, 2014
|
|
4U9H
 
 | | Ultra High Resolution Structure Of The Ni-R State Of [Nife]Hydrogenase From Desulufovibrio Vulgaris Miyazaki F | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, FE3-S4 CLUSTER, ... | | Authors: | Ogata, H, Nishikawa, K, Lubitz, W. | | Deposit date: | 2014-08-06 | | Release date: | 2015-01-21 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (0.89 Å) | | Cite: | Hydrogens detected by subatomic resolution protein crystallography in a [NiFe] hydrogenase.
Nature, 520, 2015
|
|
