4FU9
 
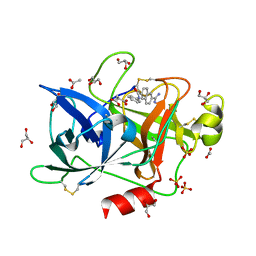 | | Crystal Structure of the Urokinase | | Descriptor: | 6-[(Z)-AMINO(IMINO)METHYL]-N-PHENYL-2-NAPHTHAMIDE, ACETATE ION, GLYCEROL, ... | | Authors: | Kang, Y.N, Stuckey, J.A, Nienaber, V, Giranda, V. | | Deposit date: | 2012-06-28 | | Release date: | 2012-08-22 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal Structure of the Urokinase
To be Published
|
|
4FUM
 
 | |
4FV0
 
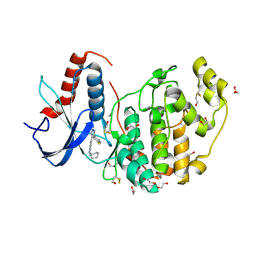 | |
4FVD
 
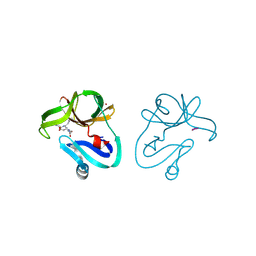 | | Crystal structure of EV71 2A proteinase C110A mutant in complex with substrate | | Descriptor: | 10-mer peptide from 2A proteinase, 2A proteinase, ZINC ION | | Authors: | Cai, Q, Muhammad, Y, Liu, W, Gao, Z, Peng, X, Cai, Y, Wu, C, Zheng, Q, Li, J, Lin, T. | | Deposit date: | 2012-06-29 | | Release date: | 2013-06-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Conformational Plasticity of 2A Proteinase from Enterovirus 71
J.Virol., 87, 2013
|
|
4FST
 
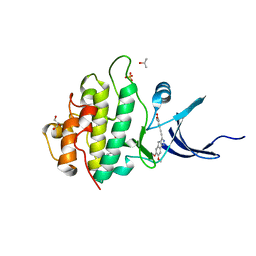 | | Crystal Structure of the CHK1 | | Descriptor: | 4-[(6,7-dimethoxy-2,4-dihydroindeno[1,2-c]pyrazol-3-yl)ethynyl]-2-methoxyphenol, GLYCEROL, ISOPROPYL ALCOHOL, ... | | Authors: | Kang, Y.N, Stuckey, J.A, Chang, P, Russell, A.J. | | Deposit date: | 2012-06-27 | | Release date: | 2012-08-22 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the CHK1
To be Published
|
|
3MEJ
 
 | | Crystal structure of putative transcriptional regulator ywtF from Bacillus subtilis, Northeast Structural Genomics Consortium Target SR736 | | Descriptor: | CHLORIDE ION, transcriptional regulator ywtF | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Mao, M, Xiao, R, Ciccosanti, C, Lee, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-03-31 | | Release date: | 2010-04-28 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (2.491 Å) | | Cite: | Northeast Structural Genomics Consortium Target SR736
To be Published
|
|
4FUF
 
 | | Crystal Structure of the Urokinase | | Descriptor: | 8-(3-bromopropoxy)-7-methoxynaphthalene-2-carboximidamide, ACETATE ION, GLYCEROL, ... | | Authors: | Kang, Y.N, Stuckey, J.A, Nienaber, V, Giranda, V. | | Deposit date: | 2012-06-28 | | Release date: | 2012-08-22 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of the Urokinase
to be published
|
|
3MGF
 
 | | Crystal Structure of Monomeric Kusabira-Orange (MKO), Orange-Emitting GFP-like Protein, at pH 7.5 | | Descriptor: | Fluorescent protein | | Authors: | Ebisawa, T, Yamamura, A, Ohtsuka, J, Kameda, Y, Hayakawa, K, Nagata, K, Tanokura, M. | | Deposit date: | 2010-04-06 | | Release date: | 2011-03-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Monomeric Kusabira-Orange (MKO), Orange-Emitting GFP-like Protein, at pH 7.5
To be Published
|
|
4FVB
 
 | | Crystal structure of EV71 2A proteinase C110A mutant | | Descriptor: | 2A proteinase, ZINC ION | | Authors: | Cai, Q, Muhammad, Y, Liu, W, Gao, Z, Peng, X, Cai, Y, Wu, C, Zheng, Q, Li, J, Lin, T. | | Deposit date: | 2012-06-29 | | Release date: | 2013-06-19 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Conformational Plasticity of 2A Proteinase from Enterovirus 71
J.Virol., 87, 2013
|
|
4FW4
 
 | | Crystal Structure of the LpxC in complex with N-[(1S,2R)-2-HYDROXY-1-(HYDROXYCARBAMOYL)PROPYL]-4-(4-PHENYLBUTA-1,3-DIYN-1-YL)BENZAMIDE inhibitor | | Descriptor: | ACETATE ION, GLYCEROL, N-[(1S,2R)-2-hydroxy-1-(hydroxycarbamoyl)propyl]-4-(4-phenylbuta-1,3-diyn-1-yl)benzamide, ... | | Authors: | Kang, Y.N, Stuckey, J.A. | | Deposit date: | 2012-06-29 | | Release date: | 2013-05-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Crystal Structure of the LpxC
to be published
|
|
4FX3
 
 | |
3MEL
 
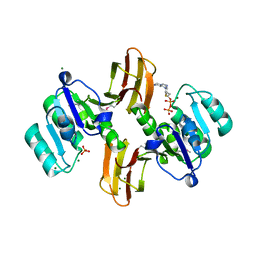 | | Crystal Structure of Thiamin pyrophosphokinase family protein from Enterococcus faecalis, Northeast Structural Genomics Consortium Target EfR150 | | Descriptor: | MAGNESIUM ION, PHOSPHATE ION, THIAMINE DIPHOSPHATE, ... | | Authors: | Kuzin, A, Abasidze, M, Seetharaman, J, Mao, M, Xiao, R, Ciccosanti, C, Foote, E.L, Maglaqui, M, Zhao, L, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-03-31 | | Release date: | 2010-05-05 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.788 Å) | | Cite: | Northeast Structural Genomics Consortium Target EfR150
To be Published
|
|
4G0H
 
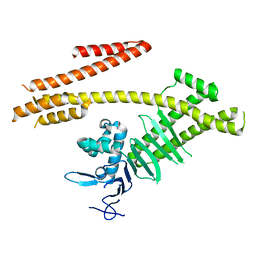 | | Crystal structure of the N-terminal domain of Helicobacter pylori CagA protein | | Descriptor: | Cytotoxicity-associated immunodominant antigen | | Authors: | Kaplan-Turkoz, B, Remaut, H, Dian, C, Louche, A, Terradot, L. | | Deposit date: | 2012-07-09 | | Release date: | 2012-08-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural insights into Helicobacter pylori oncoprotein CagA interaction with beta 1 integrin.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4G4V
 
 | |
3MJB
 
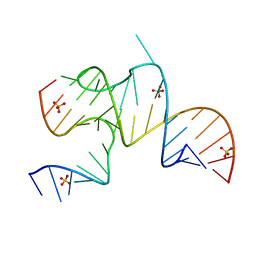 | | Cricket Paralysis Virus IGR IRES Domain 3 RNA bound to sulfate | | Descriptor: | Domain 3 of the cricket paralysis virus intergenic region IRES RNA, RNA (5'-R(P*UP*AP*AP*GP*AP*AP*AP*UP*UP*UP*AP*CP*CP*U)-3'), SULFATE ION | | Authors: | Kieft, J.S, Golden, B.L, Costantino, D.A, Chase, E. | | Deposit date: | 2010-04-12 | | Release date: | 2010-05-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Identification and characterization of anion binding sites in RNA.
Rna, 16, 2010
|
|
4G6P
 
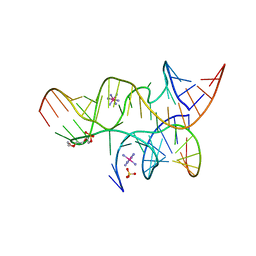 | | Minimal Hairpin Ribozyme in the Precatalytic State with A38P Variation | | Descriptor: | COBALT HEXAMMINE(III), Loop A Substrate strand, Loop A and Loop B Ribozyme strand, ... | | Authors: | Liberman, J.A, Jenkins, J.L, Krucinska, J, Wedekind, J.E. | | Deposit date: | 2012-07-19 | | Release date: | 2012-08-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.641 Å) | | Cite: | A Transition-State Interaction Shifts Nucleobase Ionization toward Neutrality To Facilitate Small Ribozyme Catalysis.
J.Am.Chem.Soc., 134, 2012
|
|
4G2E
 
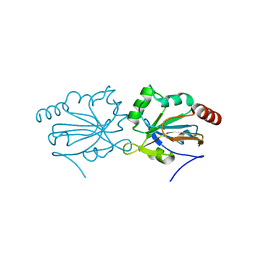 | |
4G6S
 
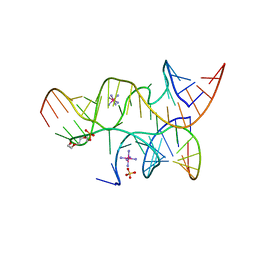 | | Minimal Hairpin Ribozyme in the Transition State with A38P Variation | | Descriptor: | COBALT HEXAMMINE(III), Loop A Substrate strand, Loop A and Loop B Ribozyme strand, ... | | Authors: | Liberman, J.A, Jenkins, J.L, Krucinska, J, Wedekind, J.E. | | Deposit date: | 2012-07-19 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | A Transition-State Interaction Shifts Nucleobase Ionization toward Neutrality To Facilitate Small Ribozyme Catalysis.
J.Am.Chem.Soc., 134, 2012
|
|
4G3T
 
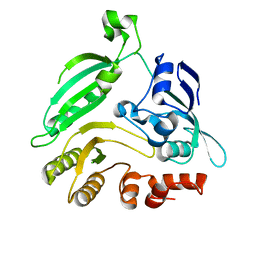 | | Mycobacterium smegmatis DprE1 - hexagonal crystal form | | Descriptor: | oxidoreductase DprE1 | | Authors: | Li, H, Jogl, G. | | Deposit date: | 2012-07-15 | | Release date: | 2012-12-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.346 Å) | | Cite: | Crystal structure of decaprenylphosphoryl-beta- D-ribose 2'-epimerase from Mycobacterium smegmatis.
Proteins, 81, 2013
|
|
4G8N
 
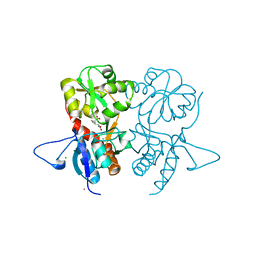 | | Crystal structure of the kainate receptor GluK3 ligand-binding domain in complex with the agonist G8M | | Descriptor: | (1S,2R)-2-[(S)-amino(carboxy)methyl]cyclobutanecarboxylic acid, CHLORIDE ION, Glutamate receptor, ... | | Authors: | Venskutonyte, R, Kastrup, J.S, Frydenvang, K, Gajhede, M. | | Deposit date: | 2012-07-23 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Pharmacological and structural characterization of conformationally restricted (S)-glutamate analogues at ionotropic glutamate receptors.
J.Struct.Biol., 180, 2012
|
|
5PBK
 
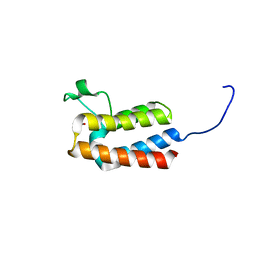 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B after initial refinement with no ligand modelled (structure 5) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
4G4F
 
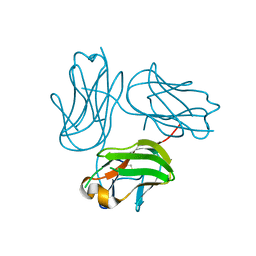 | |
5PBZ
 
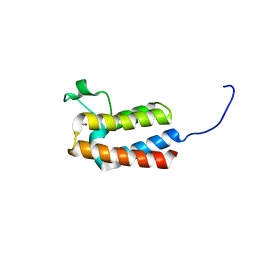 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B after initial refinement with no ligand modelled (structure 20) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
5PCD
 
 | | PanDDA analysis group deposition -- Crystal Structure of BAZ2B after initial refinement with no ligand modelled (structure 34) | | Descriptor: | 1,2-ETHANEDIOL, Bromodomain adjacent to zinc finger domain protein 2B | | Authors: | Pearce, N.M, Krojer, T, Talon, R, Bradley, A.R, Fairhead, M, Sethi, R, Wright, N, MacLean, E, Collins, P, Brandao-Neto, J, Douangamath, A, Renjie, Z, Dias, A, Vollmar, M, Ng, J, Brennan, P.E, Cox, O, Bountra, C, Arrowsmith, C.H, Edwards, A, von Delft, F. | | Deposit date: | 2017-02-03 | | Release date: | 2017-03-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | A multi-crystal method for extracting obscured crystallographic states from conventionally uninterpretable electron density.
Nat Commun, 8, 2017
|
|
3M12
 
 | |
