7CV9
 
 | | RNA methyltransferase METTL4 | | Descriptor: | GLYCEROL, Methyltransferase-like protein 2, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Luo, Q, Ma, J. | | Deposit date: | 2020-08-25 | | Release date: | 2021-09-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.455 Å) | | Cite: | Structural insights into molecular mechanism for N6-adenosine methylation by MT-A70 family methyltransferase METTL4
Nat Commun, 13, 2022
|
|
7CVA
 
 | | RNA methyltransferase METTL4 | | Descriptor: | Methyltransferase-like protein 2 | | Authors: | Luo, Q, Ma, J. | | Deposit date: | 2020-08-25 | | Release date: | 2021-09-01 | | Last modified: | 2022-10-12 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural insights into molecular mechanism for N6-adenosine methylation by MT-A70 family methyltransferase METTL4
Nat Commun, 13, 2022
|
|
7CTD
 
 | |
7CWJ
 
 | |
7CXA
 
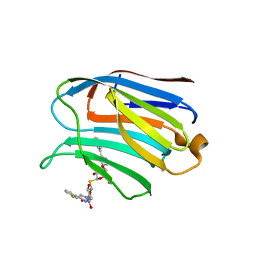 | | Structure of human Galectin-3 CRD in complex with TD-139 belonging to P31 space group. | | Descriptor: | 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-beta-D-galactopyranosyl 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-1-thio-beta-D-galactopyranoside, CHLORIDE ION, Galectin-3 | | Authors: | Kumar, A. | | Deposit date: | 2020-09-01 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Molecular mechanism of interspecies differences in the binding affinity of TD139 to Galectin-3.
Glycobiology, 31, 2021
|
|
7CTL
 
 | |
7CXC
 
 | |
7CX0
 
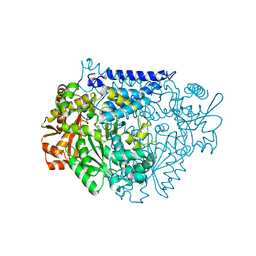 | | Crystal structure of a tyrosine decarboxylase from Enterococcus faecalis in complex with the cofactor PLP and inhibitor carbidopa | | Descriptor: | CARBIDOPA, Decarboxylase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Yu, X, Gong, M, Huang, J, Liu, W, Chen, C, Guo, R. | | Deposit date: | 2020-09-01 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Crystal structure of a tyrosine decarboxylase from Enterococcus faecalis in complex with the cofactor PLP and inhibitor carbidopa
to be published
|
|
7CWX
 
 | | Crystal structure of a tyrosine decarboxylase from Enterococcus faecalis | | Descriptor: | DI(HYDROXYETHYL)ETHER, Decarboxylase, GLYCEROL | | Authors: | Yu, X, Gong, M, Huang, J, Liu, W, Chen, C, Guo, R. | | Deposit date: | 2020-09-01 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of a tyrosine decarboxylase from Enterococcus faecalis
to be published
|
|
7CWP
 
 | |
7CWY
 
 | | Crystal structure of a tyrosine decarboxylase from Enterococcus faecalis in complex with the cofactor PLP | | Descriptor: | Decarboxylase | | Authors: | Yu, X, Gong, M, Huang, J, Liu, W, Chen, C, Guo, R. | | Deposit date: | 2020-09-01 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Crystal structure of a tyrosine decarboxylase from Enterococcus faecalis in complex with the cofactor PLP
to be published
|
|
7CV7
 
 | | RNA methyltransferase METTL4 | | Descriptor: | Methyltransferase-like protein 2, S-ADENOSYLMETHIONINE | | Authors: | Luo, Q, Ma, J. | | Deposit date: | 2020-08-25 | | Release date: | 2021-09-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural insights into molecular mechanism for N6-adenosine methylation by MT-A70 family methyltransferase METTL4
Nat Commun, 13, 2022
|
|
7CV6
 
 | | RNA methyltransferase METTL4 | | Descriptor: | 2'-O-methyladenosine 5'-(dihydrogen phosphate), Methyltransferase-like protein 2, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Luo, Q, Ma, J. | | Deposit date: | 2020-08-25 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.01 Å) | | Cite: | Structural insights into molecular mechanism for N6-adenosine methylation by MT-A70 family methyltransferase METTL4
Nat Commun, 13, 2022
|
|
7CX7
 
 | | Crystal structure of Arabinose isomerase from hybrid AI8 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, GLYCEROL, L-arabinose isomerase, ... | | Authors: | Hoang, N.K.Q, Dhanasingh, I, Cao, T.P, Sung, J.Y, Shin, S.M, Lee, D.W, Lee, S.H. | | Deposit date: | 2020-09-01 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Crystal structure of Arabinose isomerase from hyper thermophilic bacterium Thermotoga maritima (TMAI) wt
To Be Published
|
|
7CX1
 
 | | Crystal structure of a tyrosine decarboxylase from Enterococcus faecalis in complex with the cofactor PLP and inhibitor methyl-tyrosine | | Descriptor: | 4-[(2R)-2-(methylamino)propyl]phenol, Decarboxylase | | Authors: | Yu, X, Gong, M, Huang, J, Liu, W, Chen, C, Guo, R. | | Deposit date: | 2020-09-01 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Crystal structure of a tyrosine decarboxylase from Enterococcus faecalis in complex with the cofactor PLP and inhibitor methyl-tyrosine
to be published
|
|
7CXB
 
 | | Structure of mouse Galectin-3 CRD in complex with TD-139 belonging to P6522 space group. | | Descriptor: | 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-beta-D-galactopyranosyl 3-deoxy-3-[4-(3-fluorophenyl)-1H-1,2,3-triazol-1-yl]-1-thio-beta-D-galactopyranoside, CHLORIDE ION, Galectin-3 | | Authors: | Kumar, A. | | Deposit date: | 2020-09-01 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | Molecular mechanism of interspecies differences in the binding affinity of TD139 to Galectin-3.
Glycobiology, 31, 2021
|
|
7CV8
 
 | | RNA methyltransferase METTL4 | | Descriptor: | GLYCEROL, Methyltransferase-like protein 2, SINEFUNGIN | | Authors: | Luo, Q, Ma, J. | | Deposit date: | 2020-08-25 | | Release date: | 2021-09-01 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.37 Å) | | Cite: | Structural insights into molecular mechanism for N6-adenosine methylation by MT-A70 family methyltransferase METTL4
Nat Commun, 13, 2022
|
|
7CWZ
 
 | | Crystal structure of a tyrosine decarboxylase from Enterococcus faecalis K392A mutant in complex with the cofactor PLP and L-dopa | | Descriptor: | Decarboxylase, L-DOPAMINE, MAGNESIUM ION, ... | | Authors: | Yu, X, Gong, M, Huang, J, Liu, W, Chen, C, Guo, R. | | Deposit date: | 2020-09-01 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Crystal structure of a tyrosine decarboxylase from Enterococcus faecalis K392A mutant in complex with the cofactor PLP and L-dopa
to be published
|
|
7CWV
 
 | | Crystal structure of Arabinose isomerase from hyper thermophilic bacterium Thermotoga maritima (TMAI) wt | | Descriptor: | GLYCEROL, L-arabinose isomerase, MANGANESE (II) ION | | Authors: | Hoang, N.K.Q, Dhanasingh, I, Cao, T.P, Sung, J.Y, Shin, S.M, Lee, D.W, Lee, S.H. | | Deposit date: | 2020-08-31 | | Release date: | 2021-09-01 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.53 Å) | | Cite: | Crystal structure of Arabinose isomerase from hyper thermophilic bacterium Thermotoga maritima (TMAI) wt
To Be Published
|
|
7CME
 
 | | Crystal structure of human P-cadherin MEC12 (X dimer) in complex with 2-(5-chloro-2-methyl-1H-indol-3-yl)ethan-1-amine (inhibitor) | | Descriptor: | 2-(5-chloro-2-methyl-1H-indol-3-yl)ethan-1-amine, CALCIUM ION, Cadherin-3, ... | | Authors: | Senoo, A, Ito, S, Ueno, G, Nagatoishi, S, Tsumoto, K. | | Deposit date: | 2020-07-27 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Regulation of cadherin dimerization by chemical fragments as a trigger to inhibit cell adhesion
Commun Biol, 4, 2021
|
|
7CDW
 
 | |
7CMF
 
 | | Crystal structure of human P-cadherin REC12 (monomer) in complex with 2-(5-chloro-2-methyl-1H-indol-3-yl)ethan-1-amine (inhibitor) | | Descriptor: | 2-(5-chloro-2-methyl-1H-indol-3-yl)ethan-1-amine, CALCIUM ION, Cadherin-3 | | Authors: | Senoo, A, Ito, S, Ueno, G, Nagatoishi, S, Tsumoto, K. | | Deposit date: | 2020-07-27 | | Release date: | 2021-09-15 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Regulation of cadherin dimerization by chemical fragments as a trigger to inhibit cell adhesion
Commun Biol, 4, 2021
|
|
7CHL
 
 | | Crystal structure of hybrid Arabinose isomerase AI-10 | | Descriptor: | Hybrid Arabinose isomerase, MANGANESE (II) ION, SODIUM ION | | Authors: | Cao, T.P, Dhanasingh, I, Sung, J.Y, Shin, S.M, Lee, D.W, Lee, S.H. | | Deposit date: | 2020-07-06 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Crystal structure of hybrid Arabinose isomerase AI-10
To Be Published
|
|
7CH3
 
 | | Crystal structure of Arabinose isomerase from hyper thermophilic bacterium Thermotoga maritima (TMAI) triple mutant (K264A, E265A, K266A) | | Descriptor: | L-arabinose isomerase, MANGANESE (II) ION | | Authors: | Cao, T.P, Dhanasingh, I, Sung, J.Y, Shin, S.M, Lee, D.W, Lee, S.H. | | Deposit date: | 2020-07-04 | | Release date: | 2021-10-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.61 Å) | | Cite: | Crystal structure of Arabinose isomerase from hyper thermophilic bacterium Thermotoga maritima (TMAI) triple mutant (K264A, E265A, K266A)
To Be Published
|
|
7CLE
 
 | | Non-Specific Class-c acidphosphatase from Sphingobium sp. RSMS | | Descriptor: | Acid phosphatase, MAGNESIUM ION | | Authors: | Gaur, N.K, Kumar, A, Sunder, S, Mukhopadhyaya, R, Makde, R.D. | | Deposit date: | 2020-07-20 | | Release date: | 2021-11-10 | | Method: | X-RAY DIFFRACTION (2.342 Å) | | Cite: | Non-Specific Class-c acidphosphatase from Sphingobium sp. RSMS
To Be Published
|
|
