8Z9R
 
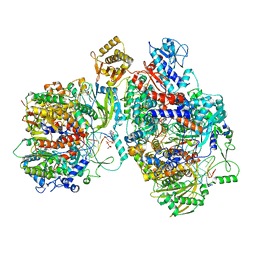 | | Cryo-EM structure of Thogoto virus polymerase in a replication elongation-reception conformation | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-D(*(ATP))-R(P*GP*CP*AP*AP*AP*AP*AP*CP*A)-3'), ... | | Authors: | Xue, L, Chang, T, Li, Z, Zhao, H, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-04-23 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.58 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses.
Nat Commun, 15, 2024
|
|
8Z85
 
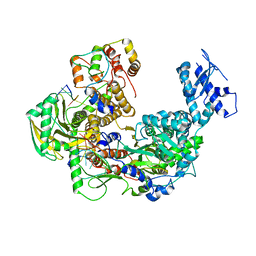 | | Cryo-EM structure of Thogoto virus polymerase in transcription pre-initiation conformation 1 | | Descriptor: | Polymerase acidic protein, Polymerase basic protein 2, RNA (5'-R(*AP*GP*AP*GP*AP*AP*AP*UP*CP*AP*AP*GP*GP*CP*AP*GP*UP*U)-3'), ... | | Authors: | Xue, L, Chang, T, Li, Z, Zhao, H, Li, M, He, J, Chen, X, Xiong, X. | | Deposit date: | 2024-04-21 | | Release date: | 2024-05-29 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Cryo-EM structures of Thogoto virus polymerase reveal unique RNA transcription and replication mechanisms among orthomyxoviruses.
Nat Commun, 15, 2024
|
|
5AMQ
 
 | |
5AMR
 
 | |
7OYG
 
 | | Dimeric form of SARS-CoV-2 RNA-dependent RNA polymerase | | Descriptor: | RNA (5'-R(P*CP*UP*AP*CP*GP*CP*AP*GP*UP*G)-3'), RNA (5'-R(P*UP*GP*CP*AP*CP*UP*GP*CP*GP*UP*AP*G)-3'), SARS-CoV-2 RNA-dependent RNA polymerase (nsp12), ... | | Authors: | Jochheim, F.A, Tegunov, D, Hillen, H.S, Schmitzova, J, Kokic, G, Dienemann, C, Cramer, P. | | Deposit date: | 2021-06-24 | | Release date: | 2021-08-25 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | The structure of a dimeric form of SARS-CoV-2 polymerase
Communications Biology, 4, 2021
|
|
3OTC
 
 | |
3OTE
 
 | |
2NVT
 
 | | RNA Polymerase II Elongation Complex in 150 mM Mg+2 with GMPCPP | | Descriptor: | 5'-D(*GP*TP*AP*CP*TP*TP*G)-3', 5'-D(P*CP*AP*AP*GP*TP*AP*CP*TP*TP*AP*CP*GP*CP*CP*TP*GP*GP*TP*CP*TP*T)-3', 5'-R(*AP*AP*GP*AP*CP*CP*AP*GP*GP*C)-3', ... | | Authors: | Wang, D, Bushnell, D.A, Westover, K.D, Kaplan, C.D, Kornberg, R.D. | | Deposit date: | 2006-11-13 | | Release date: | 2006-12-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.36 Å) | | Cite: | Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis
Cell(Cambridge,Mass.), 127, 2006
|
|
2NVY
 
 | | RNA Polymerase II form II in 150 mM Mn+2 | | Descriptor: | DNA-directed RNA polymerase II 13.6 kDa polypeptide, DNA-directed RNA polymerase II 140 kDa polypeptide, DNA-directed RNA polymerase II 45 kDa polypeptide, ... | | Authors: | Wang, D, Bushnell, D.A, Westover, K.D, Kaplan, C.D, Kornberg, R.D. | | Deposit date: | 2006-11-13 | | Release date: | 2006-12-26 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis
Cell(Cambridge,Mass.), 127, 2006
|
|
2NVZ
 
 | | RNA Polymerase II elongation complex with UTP, updated 11/2006 | | Descriptor: | 28-MER DNA template strand, 5'-D(*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*G)-3', 5'-R(*AP*UP*CP*GP*AP*GP*AP*GP*GP*A)-3', ... | | Authors: | Wang, D, Bushnell, D.A, Westover, K.D, Kaplan, C.D, Kornberg, R.D. | | Deposit date: | 2006-11-14 | | Release date: | 2006-12-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (4.3 Å) | | Cite: | Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis
Cell(Cambridge,Mass.), 127, 2006
|
|
2NVQ
 
 | | RNA Polymerase II Elongation Complex in 150 mM Mg+2 with 2'dUTP | | Descriptor: | 28-MER DNA template strand, 5'-D(*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*G)-3', 5'-R(*AP*UP*CP*GP*AP*GP*AP*GP*GP*A)-3', ... | | Authors: | Wang, D, Bushnell, D.A, Westover, K.D, Kaplan, C.D, Kornberg, R.D. | | Deposit date: | 2006-11-13 | | Release date: | 2006-12-12 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis
Cell(Cambridge,Mass.), 127, 2006
|
|
2NVX
 
 | | RNA polymerase II elongation complex in 5 mM Mg+2 with 2'-dUTP | | Descriptor: | 28-MER DNA template strand, 5'-D(*CP*TP*GP*CP*TP*TP*AP*TP*CP*GP*GP*TP*AP*G)-3', 5'-R(*AP*UP*CP*GP*AP*GP*AP*GP*GP*A)-3', ... | | Authors: | Wang, D, Bushnell, D.A, Westover, K.D, Kaplan, C.D, Kornberg, R.D. | | Deposit date: | 2006-11-13 | | Release date: | 2006-12-19 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis
Cell(Cambridge,Mass.), 127, 2006
|
|
6YYT
 
 | | Structure of replicating SARS-CoV-2 polymerase | | Descriptor: | RNA product, ZINC ION, nsp12, ... | | Authors: | Hillen, H.S, Kokic, G, Farnung, L, Dienemann, C, Tegunov, D, Cramer, P. | | Deposit date: | 2020-05-06 | | Release date: | 2020-05-13 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of replicating SARS-CoV-2 polymerase.
Nature, 584, 2020
|
|
8CI5
 
 | | Structure of the SNV L protein bound to 5' RNA | | Descriptor: | RNA (5'-R(P*AP*GP*UP*AP*GP*UP*AP*GP*AP*CP*U)-3'), RNA-directed RNA polymerase L | | Authors: | Meier, K, Thorkelsson, S.R, Durieux Trouilleton, Q, Vogel, D, Yu, D, Kosinski, J, Cusack, S, Malet, H, Grunewald, K, Quemin, E.R.J, Rosenthal, M. | | Deposit date: | 2023-02-08 | | Release date: | 2023-07-19 | | Last modified: | 2023-08-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural and functional characterization of the Sin Nombre virus L protein.
Plos Pathog., 19, 2023
|
|
6QI9
 
 | | Truncated human R2TP complex, structure 4 (ADP-empty) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, RuvB-like 1, RuvB-like 2 | | Authors: | Munoz-Hernandez, H, Rodriguez, C.F, Llorca, O. | | Deposit date: | 2019-01-18 | | Release date: | 2019-05-15 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.63 Å) | | Cite: | Structural mechanism for regulation of the AAA-ATPases RUVBL1-RUVBL2 in the R2TP co-chaperone revealed by cryo-EM.
Sci Adv, 5, 2019
|
|
6QI8
 
 | | Truncated human R2TP complex, structure 3 (ADP-filled) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, RuvB-like 1, RuvB-like 2 | | Authors: | Munoz-Hernandez, H, Rodriguez, C.F, Llorca, O. | | Deposit date: | 2019-01-18 | | Release date: | 2019-04-10 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Structural mechanism for regulation of the AAA-ATPases RUVBL1-RUVBL2 in the R2TP co-chaperone revealed by cryo-EM.
Sci Adv, 5, 2019
|
|
7ZGR
 
 | |
7ZGP
 
 | |
8FMA
 
 | | Nodavirus RNA replication proto-crown, detergent-solubliized C11 multimer | | Descriptor: | RNA-directed RNA polymerase | | Authors: | Zhan, H, Unchwaniwala, N, Rebolledo Viveros, A, Pennington, J, Horswill, M, Broadberry, R, Myers, J, den Boon, J, Grant, T, Ahlquist, P. | | Deposit date: | 2022-12-22 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Nodavirus RNA replication crown architecture reveals proto-crown precursor and viral protein A conformational switching.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8FM9
 
 | | Nodavirus RNA replication proto-crown, detergent-solubliized C12 multimer | | Descriptor: | RNA-directed RNA polymerase | | Authors: | Zhan, H, Unchwaniwala, N, Rebolledo Viveros, A, Pennington, J, Horswill, M, Broadberry, R, Myers, J, den Boon, J, Grant, T, Ahlquist, P. | | Deposit date: | 2022-12-22 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Nodavirus RNA replication crown architecture reveals proto-crown precursor and viral protein A conformational switching.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8FMB
 
 | | Nodavirus RNA replication protein A polymerase domain, local refinement | | Descriptor: | RNA-directed RNA polymerase | | Authors: | Zhan, H, Unchwaniwala, N, Rebolledo Viveros, A, Pennington, J, Horswill, M, Broadberry, R, Myers, J, den Boon, J, Grant, T, Ahlquist, P. | | Deposit date: | 2022-12-22 | | Release date: | 2023-02-01 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (6.3 Å) | | Cite: | Nodavirus RNA replication crown architecture reveals proto-crown precursor and viral protein A conformational switching.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7ZGQ
 
 | |
4YD1
 
 | | Ternary complex of human DNA Polymerase Mu with 2-nt gapped DNA substrate and an incoming nonhydrolyzable dUMPNPP | | Descriptor: | 1,2-ETHANEDIOL, 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, CHLORIDE ION, ... | | Authors: | Moon, A.F, Gosavi, R.A, Kunkel, T.A, Pedersen, L.C, Bebenek, K. | | Deposit date: | 2015-02-20 | | Release date: | 2015-08-05 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Creative template-dependent synthesis by human polymerase mu.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
7YES
 
 | |
4AU6
 
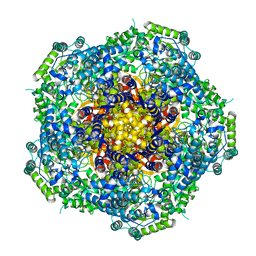 | | Location of the dsRNA-dependent polymerase, VP1, in rotavirus particles | | Descriptor: | RNA-DEPENDENT RNA POLYMERASE | | Authors: | Estrozi, L.F, Settembre, E.C, Goret, G, McClain, B, Zhang, X, Chen, J.Z, Grigorieff, N, Harrison, S.C. | | Deposit date: | 2012-05-14 | | Release date: | 2012-06-13 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Location of the Dsrna-Dependent Polymerase, Vp1, in Rotavirus Particles.
J.Mol.Biol., 425, 2013
|
|
