2RGX
 
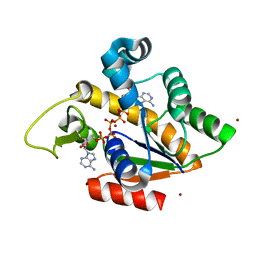 | | Crystal Structure of Adenylate Kinase from Aquifex Aeolicus in complex with Ap5A | | Descriptor: | Adenylate kinase, BIS(ADENOSINE)-5'-PENTAPHOSPHATE, ZINC ION | | Authors: | Thai, V, Wolf-Watz, M, Fenn, T, Pozharski, E, Wilson, M.A, Petsko, G.A, Kern, D. | | Deposit date: | 2007-10-05 | | Release date: | 2007-12-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Intrinsic motions along an enzymatic reaction trajectory.
Nature, 450, 2007
|
|
6GV1
 
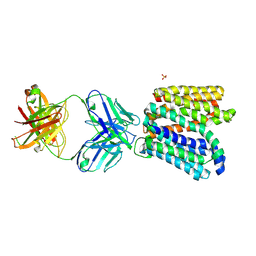 | | Crystal structure of E.coli Multidrug/H+ antiporter MdfA in outward open conformation with bound Fab fragment | | Descriptor: | Fab fragment YN1074 heavy chain, Fab fragment YN1074 light chain, Major Facilitator Superfamily multidrug/H+ antiporter MdfA from E.coli, ... | | Authors: | Nagarathinam, K, Parthier, C, Stubbs, M.T, Tanabe, M. | | Deposit date: | 2018-06-20 | | Release date: | 2018-10-03 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Outward open conformation of a Major Facilitator Superfamily multidrug/H+antiporter provides insights into switching mechanism.
Nat Commun, 9, 2018
|
|
9K8X
 
 | | Crystal structure of the calcium indicator GCaMP6s-BrUS-145 in calcium-bounded state | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, Calcium indicator GCaMP6s-BrUS-145,Calmodulin-1, ... | | Authors: | Varfolomeeva, L.A, Simonyan, T.R, Mamontova, A.V, Popov, V.O, Bogdanov, A.M, Boyko, K.M. | | Deposit date: | 2024-10-24 | | Release date: | 2024-12-11 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Calcium Indicators with Fluorescence Lifetime-Based Signal Readout: A Structure-Function Study.
Int J Mol Sci, 25, 2024
|
|
9K8W
 
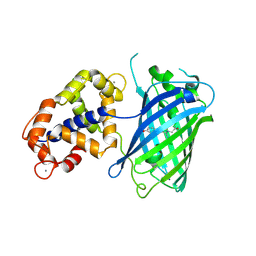 | | Crystal structure of the calcium indicator GCaMP6s-BrUS in calcium-bound state | | Descriptor: | CALCIUM ION, Calcium indicator GCaMP6s-BrUS,Calmodulin-1 | | Authors: | Varfolomeeva, L.A, Simonyan, T.R, Mamontova, A.V, Bogdanov, A.M, Popov, V.O, Boyko, K.M. | | Deposit date: | 2024-10-24 | | Release date: | 2024-12-11 | | Last modified: | 2025-01-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Calcium Indicators with Fluorescence Lifetime-Based Signal Readout: A Structure-Function Study.
Int J Mol Sci, 25, 2024
|
|
1SZR
 
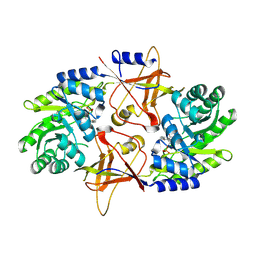 | | A Dimer interface mutant of ornithine decarboxylase reveals structure of gem diamine intermediate | | Descriptor: | N-GLYCINE-[3-HYDROXY-2-METHYL-5-PHOSPHONOOXYMETHYL-PYRIDIN-4-YL-METHANE], N~2~-({3-HYDROXY-2-METHYL-5-[(PHOSPHONOOXY)METHYL]PYRIDIN-4-YL}METHYL)-D-ORNITHINE, Ornithine decarboxylase, ... | | Authors: | Jackson, L.K, Baldwin, J, Goldsmith, E.J, Phillips, M.A. | | Deposit date: | 2004-04-06 | | Release date: | 2004-10-26 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Multiple active site conformations revealed by distant site mutation in ornithine decarboxylase
Biochemistry, 43, 2004
|
|
1TF4
 
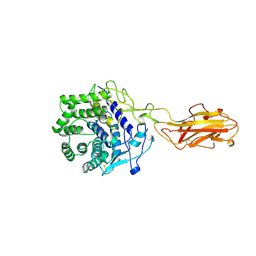 | | ENDO/EXOCELLULASE FROM THERMOMONOSPORA | | Descriptor: | CALCIUM ION, T. FUSCA ENDO/EXO-CELLULASE E4 CATALYTIC DOMAIN AND CELLULOSE-BINDING DOMAIN | | Authors: | Sakon, J, Wilson, D.B, Karplus, P.A. | | Deposit date: | 1997-05-30 | | Release date: | 1997-09-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure and mechanism of endo/exocellulase E4 from Thermomonospora fusca.
Nat.Struct.Biol., 4, 1997
|
|
1TEV
 
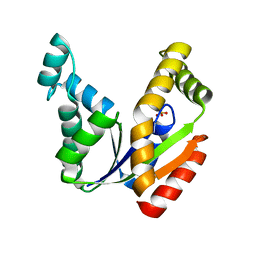 | | Crystal structure of the human UMP/CMP kinase in open conformation | | Descriptor: | SULFATE ION, UMP-CMP kinase | | Authors: | Segura-Pena, D, Sekulic, N, Ort, S, Konrad, M, Lavie, A. | | Deposit date: | 2004-05-25 | | Release date: | 2004-06-22 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Substrate-induced Conformational Changes in Human UMP/CMP Kinase.
J.Biol.Chem., 279, 2004
|
|
1SVU
 
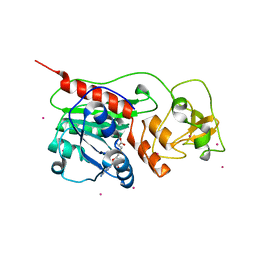 | | Structure of the Q237W mutant of HhaI DNA methyltransferase: an insight into protein-protein interactions | | Descriptor: | Modification methylase HhaI, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION, ... | | Authors: | Dong, A, Zhou, L, Zhang, X, Stickel, S, Roberts, R.J, Cheng, X. | | Deposit date: | 2004-03-30 | | Release date: | 2004-06-29 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.66 Å) | | Cite: | Structure of the Q237W mutant of HhaI DNA methyltransferase: an insight into protein-protein interactions
Biol.Chem., 385, 2004
|
|
5JUT
 
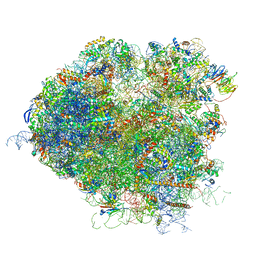 | | Saccharomyces cerevisiae 80S ribosome bound with elongation factor eEF2-GDP-sordarin and Taura Syndrome Virus IRES, Structure IV (almost non-rotated 40S subunit) | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Abeyrathne, P, Koh, C.S, Grant, T, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2016-05-10 | | Release date: | 2016-10-05 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Ensemble cryo-EM uncovers inchworm-like translocation of a viral IRES through the ribosome.
Elife, 5, 2016
|
|
8IP9
 
 | | Wheat 40S ribosome in complex with a tRNAi | | Descriptor: | 18S ribosomal RNA, 40S ribosomal protein S23, 40S ribosomal protein eS1, ... | | Authors: | Yokoyama, T, Tanaka, M, Saito, H, Nishimoto, M, Tsuda, K, Sotta, N, Shigematsu, H, Shirouzu, M, Iwasaki, S, Ito, T, Fujiwara, T. | | Deposit date: | 2023-03-14 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Boric acid intercepts 80S ribosome migration from AUG-stop by stabilizing eRF1.
Nat.Chem.Biol., 20, 2024
|
|
8IPB
 
 | | Wheat 80S ribosome pausing on AUG-Stop with cycloheximide | | Descriptor: | 18S ribosomal RNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, 40S ribosomal protein eL8, ... | | Authors: | Yokoyama, T, Tanaka, M, Saito, H, Nishimoto, M, Tsuda, K, Sotta, N, Shigematsu, H, Shirouzu, M, Iwasaki, S, Ito, T, Fujiwara, T. | | Deposit date: | 2023-03-14 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Boric acid intercepts 80S ribosome migration from AUG-stop by stabilizing eRF1.
Nat.Chem.Biol., 20, 2024
|
|
8IPA
 
 | | Wheat 80S ribosome stalled on AUG-Stop boron dependently with cycloheximide | | Descriptor: | 18S ribosomal RNA, 4-{(2R)-2-[(1S,3S,5S)-3,5-dimethyl-2-oxocyclohexyl]-2-hydroxyethyl}piperidine-2,6-dione, 40S ribosomal protein eL8, ... | | Authors: | Yokoyama, T, Tanaka, M, Saito, H, Nishimoto, M, Tsuda, K, Sotta, N, Shigematsu, H, Shirouzu, M, Iwasaki, S, Ito, T, Fujiwara, T. | | Deposit date: | 2023-03-14 | | Release date: | 2024-02-21 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Boric acid intercepts 80S ribosome migration from AUG-stop by stabilizing eRF1.
Nat.Chem.Biol., 20, 2024
|
|
5JUO
 
 | | Saccharomyces cerevisiae 80S ribosome bound with elongation factor eEF2-GDP-sordarin and Taura Syndrome Virus IRES, Structure I (fully rotated 40S subunit) | | Descriptor: | 18S ribosomal RNA, 25S ribosomal RNA, 5.8S ribosomal RNA, ... | | Authors: | Abeyrathne, P, Koh, C.S, Grant, T, Grigorieff, N, Korostelev, A.A. | | Deposit date: | 2016-05-10 | | Release date: | 2016-10-05 | | Last modified: | 2025-04-02 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Ensemble cryo-EM uncovers inchworm-like translocation of a viral IRES through the ribosome.
Elife, 5, 2016
|
|
8UKB
 
 | |
8UIZ
 
 | | In situ human P-E state 80S ribosome | | Descriptor: | 18S rRNA [Homo sapiens], 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Wei, Z, Yong, X. | | Deposit date: | 2023-10-10 | | Release date: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.43 Å) | | Cite: | In situ human P-E state 80S ribosome
To Be Published
|
|
8UJK
 
 | | In situ HHT and CHX treated A-P-Z state 80S ribosome | | Descriptor: | (3beta)-O~3~-[(2R)-2,6-dihydroxy-2-(2-methoxy-2-oxoethyl)-6-methylheptanoyl]cephalotaxine, 18S rRNA, 28S rRNA, ... | | Authors: | Wei, Z, Yong, X. | | Deposit date: | 2023-10-11 | | Release date: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (3.54 Å) | | Cite: | In situ HHT and CHX treated human A-P-Z state 80S ribosome
To Be Published
|
|
8UJ9
 
 | | In situ human P state 80S ribosome | | Descriptor: | 18S rRNA [Homo sapiens], 28S rRNA [Homo sapiens], 40S ribosomal protein S10, ... | | Authors: | Wei, Z, Yong, X. | | Deposit date: | 2023-10-11 | | Release date: | 2025-04-16 | | Method: | ELECTRON MICROSCOPY (3.64 Å) | | Cite: | In situ human P state 80S ribosome
To Be Published
|
|
7XNX
 
 | |
7XNY
 
 | |
6IP6
 
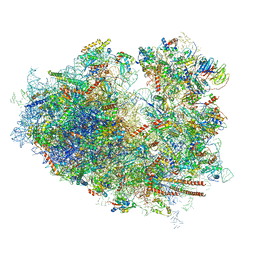 | | Cryo-EM structure of the CMV-stalled human 80S ribosome with HCV IRES (Structure iii) | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Yokoyama, T, Shigematsu, H, Shirouzu, M, Imataka, H, Ito, T. | | Deposit date: | 2018-11-02 | | Release date: | 2019-05-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | HCV IRES Captures an Actively Translating 80S Ribosome.
Mol.Cell, 74, 2019
|
|
6IP8
 
 | | Cryo-EM structure of the HCV IRES dependently initiated CMV-stalled 80S ribosome (Structure iv) | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Yokoyama, T, Shigematsu, H, Shirouzu, M, Imataka, H, Ito, T. | | Deposit date: | 2018-11-02 | | Release date: | 2019-05-29 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | HCV IRES Captures an Actively Translating 80S Ribosome.
Mol.Cell, 74, 2019
|
|
9BDP
 
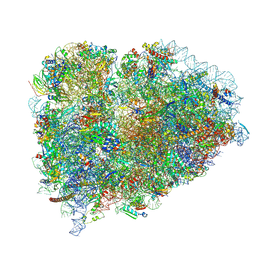 | |
9BDN
 
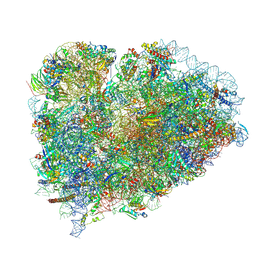 | |
7NRD
 
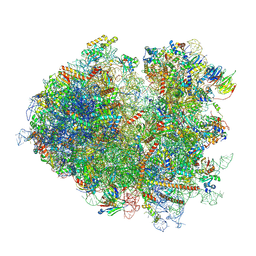 | | Structure of the yeast Gcn1 bound to a colliding stalled 80S ribosome with MBF1, A/P-tRNA and P/E-tRNA | | Descriptor: | 25S rRNA (3184-MER), 40S ribosomal protein S0-A, 40S ribosomal protein S1-A, ... | | Authors: | Pochopien, A.A, Beckert, B, Wilson, D.N. | | Deposit date: | 2021-03-03 | | Release date: | 2021-04-14 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (4.36 Å) | | Cite: | Structure of Gcn1 bound to stalled and colliding 80S ribosomes.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7NRC
 
 | | Structure of the yeast Gcn1 bound to a leading stalled 80S ribosome with Rbg2, Gir2, A- and P-tRNA and eIF5A | | Descriptor: | 18S rRNA (1771-MER), 25S rRNA (3184-MER), 40S ribosomal protein S0-A, ... | | Authors: | Pochopien, A.A, Beckert, B, Wilson, D.N. | | Deposit date: | 2021-03-03 | | Release date: | 2021-05-05 | | Last modified: | 2025-04-09 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structure of Gcn1 bound to stalled and colliding 80S ribosomes.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
