3UQF
 
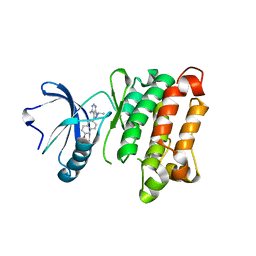 | | c-SRC kinase domain in complex with BKI RM-1-89 | | Descriptor: | 3-(6-ethoxynaphthalen-2-yl)-1-(propan-2-yl)-1H-pyrazolo[3,4-d]pyrimidin-4-amine, Proto-oncogene tyrosine-protein kinase Src | | Authors: | Merritt, E.A, Larson, E.T. | | Deposit date: | 2011-11-20 | | Release date: | 2012-03-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Multiple Determinants for Selective Inhibition of Apicomplexan Calcium-Dependent Protein Kinase CDPK1.
J.Med.Chem., 55, 2012
|
|
1UPJ
 
 | |
1I72
 
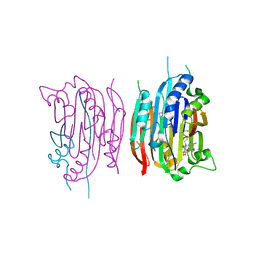 | | HUMAN S-ADENOSYLMETHIONINE DECARBOXYLASE WITH COVALENTLY BOUND PYRUVOYL GROUP AND COVALENTLY BOUND 5'-DEOXY-5'-[N-METHYL-N-(2-AMINOOXYETHYL) AMINO]ADENOSINE | | Descriptor: | 1,4-DIAMINOBUTANE, 5'-DEOXY-5'-[N-METHYL-N-(2-AMINOOXYETHYL) AMINO]ADENOSINE, S-ADENOSYLMETHIONINE DECARBOXYLASE ALPHA CHAIN, ... | | Authors: | Tolbert, W.D, Ekstrom, J.L, Mathews, I.I, Secrist III, J.A, Pegg, A.E, Ealick, S.E. | | Deposit date: | 2001-03-07 | | Release date: | 2001-08-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The structural basis for substrate specificity and inhibition of human S-adenosylmethionine decarboxylase.
Biochemistry, 40, 2001
|
|
1QO5
 
 | |
2IN6
 
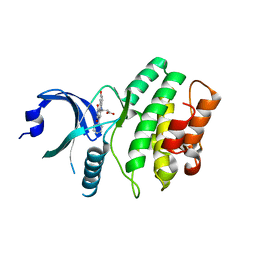 | | Wee1 kinase complex with inhibitor PD311839 | | Descriptor: | 3-(9-HYDROXY-1,3-DIOXO-4-PHENYL-2,3-DIHYDROPYRROLO[3,4-C]CARBAZOL-6(1H)-YL)PROPANOIC ACID, Wee1-like protein kinase | | Authors: | Squire, C.J, Dickson, J.M, Ivanovic, I, Baker, E.N. | | Deposit date: | 2006-10-05 | | Release date: | 2007-09-18 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Synthesis and structure-activity relationships of N-6 substituted analogues of 9-hydroxy-4-phenylpyrrolo[3,4-c]carbazole-1,3(2H,6H)-diones as inhibitors of Wee1 and Chk1 checkpoint kinases.
Eur.J.Med.Chem., 43, 2008
|
|
3VSB
 
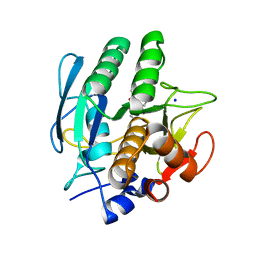 | | SUBTILISIN CARLSBERG D-NAPHTHYL-1-ACETAMIDO BORONIC ACID INHIBITOR COMPLEX | | Descriptor: | SODIUM ION, SUBTILISIN CARLSBERG, TYPE VIII | | Authors: | Stoll, V.S, Eger, B.T, Hynes, R.C, Martichonok, V, Jones, J.B, Pai, E.F. | | Deposit date: | 1997-09-25 | | Release date: | 1998-03-25 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Differences in binding modes of enantiomers of 1-acetamido boronic acid based protease inhibitors: crystal structures of gamma-chymotrypsin and subtilisin Carlsberg complexes.
Biochemistry, 37, 1998
|
|
1YIF
 
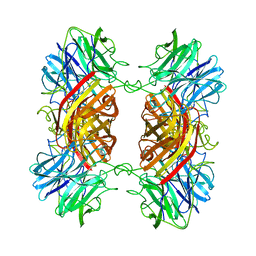 | | CRYSTAL STRUCTURE OF beta-1,4-xylosidase FROM BACILLUS SUBTILIS, NEW YORK STRUCTURAL GENOMICS CONSORTIUM | | Descriptor: | BETA-1,4-XYLOSIDASE | | Authors: | Patskovsky, Y, Almo, S.C, Burley, S.K, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2005-01-11 | | Release date: | 2005-01-18 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | CRYSTAL STRUCTURE OF beta-1,4-xylosidase FROM BACILLUS SUBTILIS
To be Published
|
|
1ICH
 
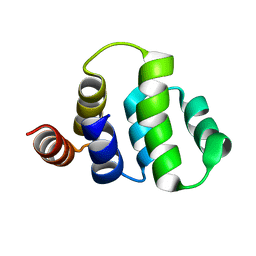 | | SOLUTION STRUCTURE OF THE TUMOR NECROSIS FACTOR RECEPTOR-1 DEATH DOMAIN | | Descriptor: | TUMOR NECROSIS FACTOR RECEPTOR-1 | | Authors: | Sukits, S.F, Lin, L.-L, Malakian, K, Powers, R, Xu, G.-Y. | | Deposit date: | 2001-04-01 | | Release date: | 2002-04-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the tumor necrosis factor receptor-1 death domain.
J.Mol.Biol., 310, 2001
|
|
1I79
 
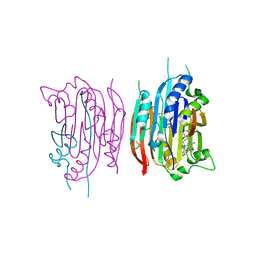 | | HUMAN S-ADENOSYLMETHIONINE DECARBOXYLASE WITH COVALENTLY BOUND PYRUVOYL GROUP AND COVALENTLY BOUND 5'-DEOXY-5'-[(3-HYDRAZINOPROPYL)METHYLAMINO]ADENOSINE | | Descriptor: | 1,4-DIAMINOBUTANE, 5'-DEOXY-5'-[(3-HYDRAZINOPROPYL)METHYLAMINO]ADENOSINE, S-ADENOSYLMETHIONINE DECARBOXYLASE ALPHA CHAIN, ... | | Authors: | Tolbert, W.D, Ekstrom, J.L, Mathews, I.I, Secrist III, J.A, Pegg, A.E, Ealick, S.E. | | Deposit date: | 2001-03-08 | | Release date: | 2001-08-22 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The structural basis for substrate specificity and inhibition of human S-adenosylmethionine decarboxylase.
Biochemistry, 40, 2001
|
|
1W6M
 
 | | X-RAY CRYSTAL STRUCTURE OF C2S HUMAN GALECTIN-1 COMPLEXED WITH GALACTOSE | | Descriptor: | BETA-MERCAPTOETHANOL, GALECTIN-1, SULFATE ION, ... | | Authors: | Lopez-Lucendo, M.I.F, Gabius, H.J, Romero, A. | | Deposit date: | 2004-08-19 | | Release date: | 2004-10-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Growth-Regulatory Human Galectin-1: Crystallographic Characterisation of the Structural Changes Induced by Single-Site Mutations and Their Impact on the Thermodynamics of Ligand Binding.
J.Mol.Biol., 343, 2004
|
|
1DAE
 
 | | DETHIOBIOTIN SYNTHETASE COMPLEXED WITH 3-(1-AMINOETHYL) NONANEDIOIC ACID | | Descriptor: | 3-(1-AMINOETHYL)NONANEDIOIC ACID, DETHIOBIOTIN SYNTHETASE | | Authors: | Huang, W, Jia, J, Schneider, G, Lindqvist, Y. | | Deposit date: | 1995-05-08 | | Release date: | 1996-06-20 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Mechanism of an ATP-dependent carboxylase, dethiobiotin synthetase, based on crystallographic studies of complexes with substrates and a reaction intermediate.
Biochemistry, 34, 1995
|
|
7XB0
 
 | | Crystal structure of Omicron BA.2 RBD complexed with hACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | Li, L, Liao, H, Meng, Y, Li, W. | | Deposit date: | 2022-03-19 | | Release date: | 2022-07-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of human ACE2 higher binding affinity to currently circulating Omicron SARS-CoV-2 sub-variants BA.2 and BA.1.1.
Cell, 185, 2022
|
|
3D0E
 
 | | Crystal structure of human Akt2 in complex with GSK690693 | | Descriptor: | 4-{2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-[(3S)-piperidin-3-ylmethoxy]-1H-imidazo[4,5-c]pyridin-4-yl}-2-methylbut-3 -yn-2-ol, RAC-beta serine/threonine-protein kinase | | Authors: | Concha, N.O, Smallwood, A. | | Deposit date: | 2008-05-01 | | Release date: | 2008-10-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of 4-(2-(4-amino-1,2,5-oxadiazol-3-yl)-1-ethyl-7-{[(3S)-3-piperidinylmethyl]oxy}-1H-imidazo[4,5-c]pyridin-4-yl)-2-methyl-3-butyn-2-ol (GSK690693), a novel inhibitor of AKT kinase.
J.Med.Chem., 51, 2008
|
|
1IFU
 
 | | RICIN A-CHAIN (RECOMBINANT) COMPLEX WITH FORMYCIN | | Descriptor: | (1S)-1-(7-amino-1H-pyrazolo[4,3-d]pyrimidin-3-yl)-1,4-anhydro-D-ribitol, RICIN | | Authors: | Weston, S.A, Tucker, A.D, Thatcher, D.R, Derbyshire, D.J, Pauptit, R.A. | | Deposit date: | 1996-07-05 | | Release date: | 1998-01-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | X-ray structure of recombinant ricin A-chain at 1.8 A resolution.
J.Mol.Biol., 244, 1994
|
|
2END
 
 | | CRYSTAL STRUCTURE OF A PYRIMIDINE DIMER SPECIFIC EXCISION REPAIR ENZYME FROM BACTERIOPHAGE T4: REFINEMENT AT 1.45 ANGSTROMS AND X-RAY ANALYSIS OF THE THREE ACTIVE SITE MUTANTS | | Descriptor: | ENDONUCLEASE V | | Authors: | Vassylyev, D.G, Ariyoshi, M, Matsumoto, O, Katayanagi, K, Ohtsuka, E, Morikawa, K. | | Deposit date: | 1994-08-08 | | Release date: | 1994-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of a pyrimidine dimer-specific excision repair enzyme from bacteriophage T4: refinement at 1.45 A and X-ray analysis of the three active site mutants.
J.Mol.Biol., 249, 1995
|
|
4ASY
 
 | | Pseudomonas aeruginosa RmlA in complex with allosteric inhibitor | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, GLUCOSE-1-PHOSPHATE THYMIDYLYLTRANSFERASE, ... | | Authors: | Alphey, M.S, Pirrie, L, Torrie, L.S, Gardiner, M, Sarkar, A, Brenk, R, Westwood, N.J, Gray, D, Naismith, J.H. | | Deposit date: | 2012-05-03 | | Release date: | 2012-10-31 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Allosteric competitive inhibitors of the glucose-1-phosphate thymidylyltransferase (RmlA) from Pseudomonas aeruginosa.
ACS Chem. Biol., 8, 2013
|
|
1DWD
 
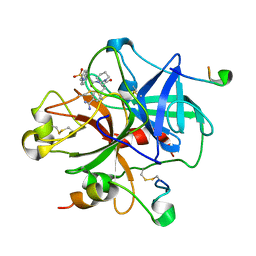 | |
1ZPI
 
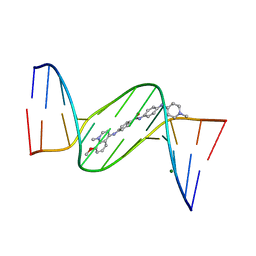 | | Crystal structure analysis of the minor groove binding quinolinium quaternary salt SN 8224 complexed with CGCGAATTCGCG | | Descriptor: | 5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3', 8-METHOXY-1-METHYL-4-(4-(4-(1-METHYLPYRIDINIUM-4-YLAMINO)PHENYLCARBAMOYL)PHENYLAMINO)QUINOLINIUM, MAGNESIUM ION | | Authors: | Adams, A, Leong, C, Denny, W.A, Guss, J.M. | | Deposit date: | 2005-05-16 | | Release date: | 2005-10-11 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of two minor-groove-binding quinolinium quaternary salts complexed with d(CGCGAATTCGCG)(2) at 1.6 and 1.8 Angstrom resolution.
Acta Crystallogr.,Sect.D, 61, 2005
|
|
1IJV
 
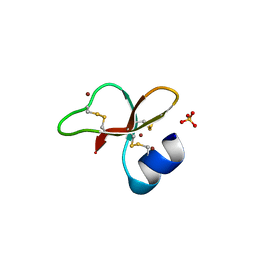 | | HUMAN BETA-DEFENSIN-1 | | Descriptor: | BROMIDE ION, Beta-defensin 1, POTASSIUM ION, ... | | Authors: | Hoover, D.M, Lubkowski, J. | | Deposit date: | 2001-04-30 | | Release date: | 2001-10-24 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | The structure of human beta-defensin-1: new insights into structural properties of beta-defensins.
J.Biol.Chem., 276, 2001
|
|
1ZA8
 
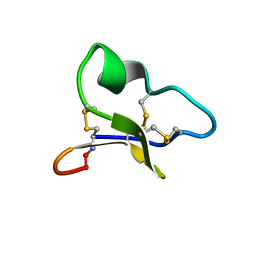 | | NMR solution structure of a leaf-specific-expressed cyclotide vhl-1 | | Descriptor: | vhl-1 | | Authors: | Chen, B, Colgrave, M.L, Daly, N.L, Rosengren, K.J, Gustafson, K.R, Craik, D.J. | | Deposit date: | 2005-04-05 | | Release date: | 2005-04-12 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | Isolation and characterization of novel cyclotides from Viola hederaceae: solution structure and anti-HIV activity of vhl-1, a leaf-specific expressed cyclotide.
J.Biol.Chem., 280, 2005
|
|
4FIV
 
 | | FIV PROTEASE COMPLEXED WITH AN INHIBITOR LP-130 | | Descriptor: | 4-[2-(2-ACETYLAMINO-3-NAPHTALEN-1-YL-PROPIONYLAMINO)-4-METHYL-PENTANOYLAMINO]-3-HYDROXY-6-METHYL-HEPTANOIC ACID [1-(1-CARBAMOYL-2-NAPHTHALEN-1-YL-ETHYLCARBAMOYL)-PROPYL]-AMIDE, FELINE IMMUNODEFICIENCY VIRUS PROTEASE | | Authors: | Kervinen, J, Lubkowski, J, Zdanov, A, Wlodawer, A, Gustchina, A. | | Deposit date: | 1998-07-15 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Toward a universal inhibitor of retroviral proteases: comparative analysis of the interactions of LP-130 complexed with proteases from HIV-1, FIV, and EIAV.
Protein Sci., 7, 1998
|
|
2OC4
 
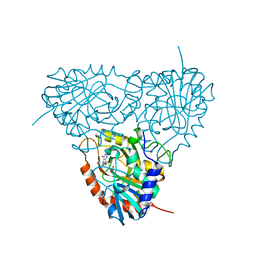 | | Crystal structure of human purine nucleoside phosphorylase mutant H257D with Imm-H | | Descriptor: | 1,4-DIDEOXY-4-AZA-1-(S)-(9-DEAZAHYPOXANTHIN-9-YL)-D-RIBITOL, PHOSPHATE ION, Purine nucleoside phosphorylase | | Authors: | Rinaldo-Matthis, A, Almo, S.C, Schramm, V.L. | | Deposit date: | 2006-12-20 | | Release date: | 2007-05-22 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.592 Å) | | Cite: | Neighboring Group Participation in the Transition State of Human Purine Nucleoside Phosphorylase
Biochemistry, 46, 2007
|
|
7XSJ
 
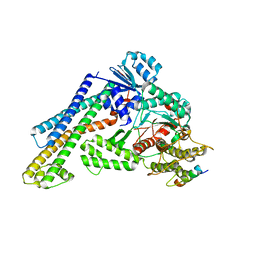 | | The structure of the Mint1/Munc18-1/syntaxin-1 complex | | Descriptor: | Amyloid-beta A4 precursor protein-binding family A member 1, Syntaxin-1A, Syntaxin-binding protein 1 | | Authors: | Feng, W, Li, W. | | Deposit date: | 2022-05-14 | | Release date: | 2022-11-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A non-canonical target-binding site in Munc18-1 domain 3b for assembling the Mint1-Munc18-1-syntaxin-1 complex.
Structure, 31, 2023
|
|
3N98
 
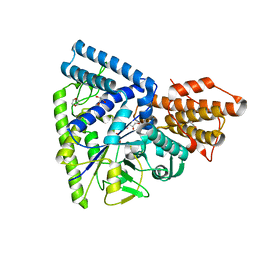 | | Crystal structure of TK1436, a GH57 branching enzyme from hyperthermophilic archaeon Thermococcus kodakaraensis, in complex with glucose and additives | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Santos, C.R, Tonoli, C.C.C, Trindade, D.M, Betzel, C, Takata, H, Kuriki, T, Kanai, T, Imanaka, T, Arni, R.K, Murakami, M.T. | | Deposit date: | 2010-05-28 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Structural basis for branching-enzyme activity of glycoside hydrolase family 57: Structure and stability studies of a novel branching enzyme from the hyperthermophilic archaeon Thermococcus Kodakaraensis KOD1.
Proteins, 79, 2011
|
|
3FEH
 
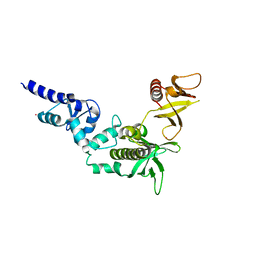 | | Crystal structure of full length centaurin alpha-1 | | Descriptor: | Centaurin-alpha-1, UNKNOWN ATOM OR ION, ZINC ION | | Authors: | Shen, L, Tong, Y, Tempel, W, MacKenzie, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J, Bochkarev, A, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2008-11-29 | | Release date: | 2008-12-23 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Phosphorylation-independent dual-site binding of the FHA domain of KIF13 mediates phosphoinositide transport via centaurin {alpha}1.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
