8SVU
 
 | | Crystal structure of the L428V mutant of pregnane X receptor ligand binding domain in apo form | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 1 group I member 2, Nuclear receptor coactivator 1 fusion protein,Nuclear receptor coactivator 1 | | Authors: | Garcia-Maldonado, E, Huber, A.D, Nithianantham, S, Miller, D.J, Chen, T. | | Deposit date: | 2023-05-17 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Chemical manipulation of an activation/inhibition switch in the nuclear receptor PXR.
Nat Commun, 15, 2024
|
|
8SVT
 
 | | Crystal structure of pregnane X receptor ligand binding domain in complex with SJPYT-331 | | Descriptor: | DIMETHYL SULFOXIDE, Nuclear receptor subfamily 1 group I member 2, Nuclear receptor coactivator 1 fusion protein,Nuclear receptor coactivator 1, ... | | Authors: | Garcia-Maldonado, E, Huber, A.D, Nithianantham, S, Miller, D.J, Chen, T. | | Deposit date: | 2023-05-17 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.39 Å) | | Cite: | Chemical manipulation of an activation/inhibition switch in the nuclear receptor PXR.
Nat Commun, 15, 2024
|
|
6UUW
 
 | | Structure of human ATP citrate lyase E599Q mutant in complex with Mg2+, citrate, ATP and CoA | | Descriptor: | (2S)-2-hydroxy-2-[2-oxo-2-(phosphonooxy)ethyl]butanedioic acid, ADENOSINE-5'-DIPHOSPHATE, ATP-citrate synthase, ... | | Authors: | Wei, X, Marmorstein, R. | | Deposit date: | 2019-11-01 | | Release date: | 2019-12-25 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (2.85 Å) | | Cite: | Molecular basis for acetyl-CoA production by ATP-citrate lyase
Nat.Struct.Mol.Biol., 27, 2020
|
|
6V88
 
 | |
1X8S
 
 | |
5C0K
 
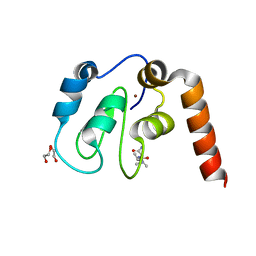 | | Fragment-Based Drug Discovery Targeting Inhibitor of Apoptosis Proteins: Compound 3 | | Descriptor: | (2S)-1-{[(2S)-1-amino-3-methyl-1-oxobutan-2-yl]amino}-1-oxopropan-2-aminium, 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase XIAP, ... | | Authors: | Chessari, G, Buck, I.M, Day, J.E.H, Day, P.J, Iqbal, A, Johnson, C.N, Lewis, E.J, Martins, V, Miller, D, Reader, M, Rees, D.C, Rich, S.J, Tamanini, E, Vitorino, M, Ward, G.A, Williams, P.A, Williams, G, Wilsher, N.E, Woolford, A.J.-A. | | Deposit date: | 2015-06-12 | | Release date: | 2015-08-12 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Fragment-Based Drug Discovery Targeting Inhibitor of Apoptosis Proteins: Discovery of a Non-Alanine Lead Series with Dual Activity Against cIAP1 and XIAP.
J.Med.Chem., 58, 2015
|
|
5FP5
 
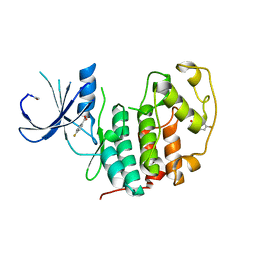 | | Structure of cyclin-dependent kinase 2 with small-molecule ligand 4- fluorobenzoic acid (AT222) in an alternate binding site. | | Descriptor: | 4-fluorobenzoic acid, ACETYL GROUP, CYCLIN-DEPENDENT KINASE 2 | | Authors: | Jhoti, H, Ludlow, R.F, O'Reilly, M, Saini, H.K, Tickle, I.J, Verdonk, M. | | Deposit date: | 2015-11-27 | | Release date: | 2015-12-09 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Detection of Secondary Binding Sites in Proteins Using Fragment Screening.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
6W9V
 
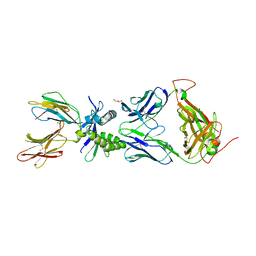 | |
7BVW
 
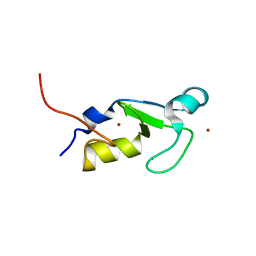 | | Crystal structure of the RING-H2 domain of Arabidopsis RMR1 | | Descriptor: | AT5G66160 protein, SODIUM ION, ZINC ION | | Authors: | Chen, S, Wong, K.B. | | Deposit date: | 2020-04-12 | | Release date: | 2021-04-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The RING-finger of AtRMR1 (Arabidopsis receptor-homology-transmembrane-RING-H2 sorting receptor 1) is an E3 ligase that mediate its trafficking
To Be Published
|
|
8CUY
 
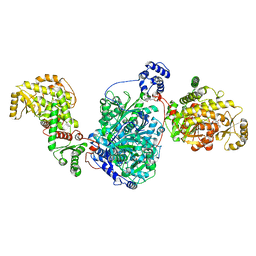 | | ACP1-KS-AT domains of mycobacterial Pks13 | | Descriptor: | 4'-PHOSPHOPANTETHEINE, Polyketide synthase PKS13, UNKNOWN LIGAND | | Authors: | Kim, S.K, Dickinson, M.S, Finer-Moore, J.S, Rosenberg, O.S, Stroud, R.M. | | Deposit date: | 2022-05-17 | | Release date: | 2023-02-15 | | Last modified: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (2.4 Å) | | Cite: | Structure and dynamics of the essential endogenous mycobacterial polyketide synthase Pks13.
Nat.Struct.Mol.Biol., 30, 2023
|
|
7PMK
 
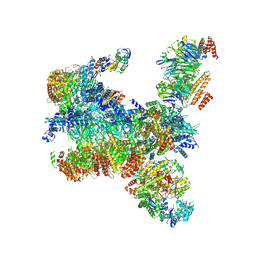 | | S. cerevisiae replisome-SCF(Dia2) complex bound to double-stranded DNA (conformation I) | | Descriptor: | Cell division control protein 45,Cell division control protein 45, Chromosome segregation in meiosis protein 3, DNA helicase, ... | | Authors: | Jenkyn-Bedford, M, Yeeles, J.T.P, Deegan, T.D. | | Deposit date: | 2021-09-02 | | Release date: | 2021-11-10 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | A conserved mechanism for regulating replisome disassembly in eukaryotes.
Nature, 600, 2021
|
|
1ERR
 
 | |
6BZ2
 
 | | Crystal structure of wild-type HIV-1 protease with a novel HIV-1 inhibitor GRL-14213A of 6-5-5-ring fused crown-like tetrahydropyranofuran as the P2-ligand, a cyclopropylaminobenzothiazole as the P2'-ligand and 3,5-difluorophenylmethyl as the P1-ligand | | Descriptor: | (3S,3aR,5R,7aS,8S)-hexahydro-4H-3,5-methanofuro[2,3-b]pyran-8-yl [(2S,3R)-4-[{[2-(cyclopropylamino)-1,3-benzothiazol-6-yl]sulfonyl}(2-methylpropyl)amino]-1-(3,5-difluorophenyl)-3-hydroxybutan-2-yl]carbamate, ACETATE ION, CHLORIDE ION, ... | | Authors: | Wang, Y.-F, Agniswamy, J, Weber, I.T. | | Deposit date: | 2017-12-22 | | Release date: | 2018-02-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Design of Highly Potent, Dual-Acting and Central-Nervous-System-Penetrating HIV-1 Protease Inhibitors with Excellent Potency against Multidrug-Resistant HIV-1 Variants.
ChemMedChem, 13, 2018
|
|
8D52
 
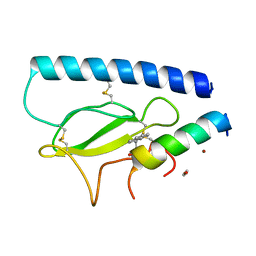 | | Parathyroid hormone 1 receptor extracellular domain complexed with a peptide ligand containing (2-naphthyl)-beta-3-homoalanine | | Descriptor: | 1,2-ETHANEDIOL, PTHrP[1-36], Parathyroid hormone/parathyroid hormone-related peptide receptor, ... | | Authors: | Yu, Z, Kreitler, D.F, Gellman, S.H. | | Deposit date: | 2022-06-03 | | Release date: | 2023-06-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Harnessing Aromatic-Histidine Interactions through Synergistic Backbone Extension and Side Chain Modification.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
8D51
 
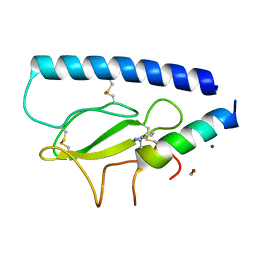 | | Parathyroid hormone 1 receptor extracellular domain complexed with a peptide ligand containing beta-3-homotryptophan | | Descriptor: | 1,2-ETHANEDIOL, PTHrP[1-36], Parathyroid hormone/parathyroid hormone-related peptide receptor, ... | | Authors: | Yu, Z, Kreitler, D.F, Gellman, S.H. | | Deposit date: | 2022-06-03 | | Release date: | 2023-06-14 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Harnessing Aromatic-Histidine Interactions through Synergistic Backbone Extension and Side Chain Modification.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
6GGP
 
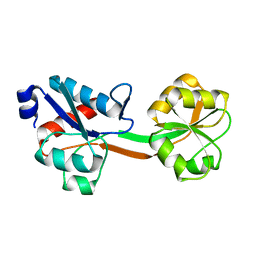 | | Structure of the ligand-free form of truncated ArgBP (residues 20-233) from T. maritima | | Descriptor: | Amino acid ABC transporter, periplasmic amino acid-binding protein | | Authors: | Smaldone, G, Berisio, R, Balasco, N, D'Auria, S, Vitagliano, L, Ruggiero, A. | | Deposit date: | 2018-05-03 | | Release date: | 2018-06-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.03 Å) | | Cite: | Domain swapping dissection in Thermotoga maritima arginine binding protein: How structural flexibility may compensate destabilization.
Biochim. Biophys. Acta, 1866, 2018
|
|
6GL4
 
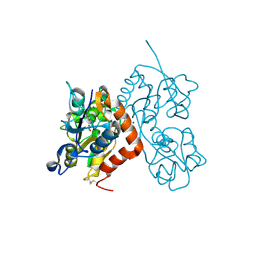 | | Structure of GluA2o ligand-binding domain (S1S2J) in complex with glutamate and sodium bromide at 1.95 A resolution | | Descriptor: | ACETATE ION, BROMIDE ION, GLUTAMIC ACID, ... | | Authors: | Venskutonyte, R, Frydenvang, K, Kastrup, J.S. | | Deposit date: | 2018-05-22 | | Release date: | 2019-05-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.948 Å) | | Cite: | Nanoscale Mobility of the Apo State and TARP Stoichiometry Dictate the Gating Behavior of Alternatively Spliced AMPA Receptors.
Neuron, 102, 2019
|
|
1R17
 
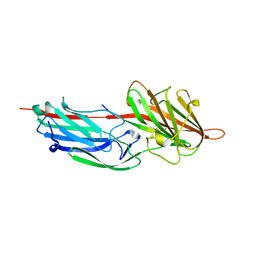 | | Crystal Structure Analysis of S.epidermidis adhesin SdrG binding to Fibrinogen (adhesin-ligand complex) | | Descriptor: | CALCIUM ION, fibrinogen-binding protein SdrG, fibrinopeptide B | | Authors: | Ponnuraj, K, Bowden, M.G, Davis, S, Gurusiddappa, S, Moore, D, Choe, D, Xu, Y, Hook, M, Narayana, S.V.L. | | Deposit date: | 2003-09-23 | | Release date: | 2003-10-28 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | A "dock, lock and latch" Structural Model for a Staphylococcal Adhesin Binding to Fibrinogen
Cell(Cambridge,Mass.), 115, 2003
|
|
5O25
 
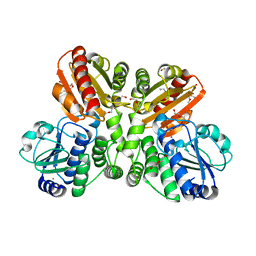 | | Structure of wildtype T.maritima PDE (TM1595) in ligand-free state | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, MANGANESE (II) ION, ... | | Authors: | Witte, G, Drexler, D, Mueller, M. | | Deposit date: | 2017-05-19 | | Release date: | 2017-10-25 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural and Biophysical Analysis of the Soluble DHH/DHHA1-Type Phosphodiesterase TM1595 from Thermotoga maritima.
Structure, 25, 2017
|
|
4N6D
 
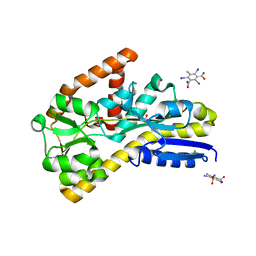 | | Crystal structure of a TRAP periplasmic solute binding protein from Desulfovibrio salexigens DSM2638 (Desal_3247), Target EFI-510112, phased with I3C, open complex, C-terminus of symmetry mate bound in ligand binding site | | Descriptor: | 1,2-ETHANEDIOL, 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, CHLORIDE ION, ... | | Authors: | Vetting, M.W, Al Obaidi, N.F, Stead, M, Attonito, J.D, Scott Glenn, A, Chowdhury, S, Evans, B, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-10-11 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.701 Å) | | Cite: | Experimental strategies for functional annotation and metabolism discovery: targeted screening of solute binding proteins and unbiased panning of metabolomes.
Biochemistry, 54, 2015
|
|
1D5F
 
 | | STRUCTURE OF E6AP: INSIGHTS INTO UBIQUITINATION PATHWAY | | Descriptor: | E6AP HECT CATALYTIC DOMAIN, E3 LIGASE | | Authors: | Huang, L, Kinnucan, E, Wang, G, Beaudenon, S, Howley, P.M, Huibregtse, J.M, Pavletich, N.P. | | Deposit date: | 1999-10-07 | | Release date: | 1999-11-17 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of an E6AP-UbcH7 complex: insights into ubiquitination by the E2-E3 enzyme cascade.
Science, 286, 1999
|
|
3EQS
 
 | |
4KNG
 
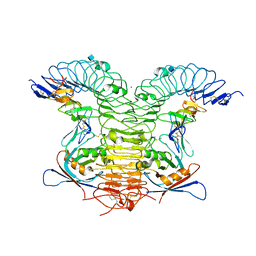 | | Crystal structure of human LGR5-RSPO1-RNF43 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, E3 ubiquitin-protein ligase RNF43, Leucine-rich repeat-containing G-protein coupled receptor 5, ... | | Authors: | Chen, P.H, He, X. | | Deposit date: | 2013-05-09 | | Release date: | 2013-06-19 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The structural basis of R-spondin recognition by LGR5 and RNF43.
Genes Dev., 27, 2013
|
|
1IF9
 
 | | Carbonic Anhydrase II Complexed With N-[2-(1H-Indol-5-yl)-butyl]-4-sulfamoyl-benzamide | | Descriptor: | CARBONIC ANHYDRASE II, MERCURY (II) ION, N-[2-(1H-INDOL-5-YL)-BUTYL]-4-SULFAMOYL-BENZAMIDE, ... | | Authors: | Grzybowski, B.A, Ishchenko, A.V, Kim, C.-Y, Topalov, G, Chapman, R, Christianson, D.W, Whitesides, G.M, Shakhnovich, E.I. | | Deposit date: | 2001-04-12 | | Release date: | 2001-05-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Combinatorial computational method gives new picomolar ligands for a known enzyme.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
1IF7
 
 | | Carbonic Anhydrase II Complexed With (R)-N-(3-Indol-1-yl-2-methyl-propyl)-4-sulfamoyl-benzamide | | Descriptor: | (R)-N-(3-INDOL-1-YL-2-METHYL-PROPYL)-4-SULFAMOYL-BENZAMIDE, CARBONIC ANHYDRASE II, MERCURY (II) ION, ... | | Authors: | Grzybowski, B.A, Ishchenko, A.V, Kim, C.-Y, Topalov, G, Chapman, R, Christianson, D.W, Whitesides, G.M, Shakhnovich, E.I. | | Deposit date: | 2001-04-12 | | Release date: | 2001-05-02 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Combinatorial computational method gives new picomolar ligands for a known enzyme.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
