105D
 
 | |
106D
 
 | |
3MMC
 
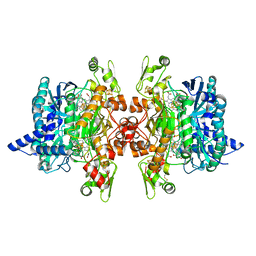 | | Structure of the dissimilatory sulfite reductase from Archaeoglobus fulgidus | | Descriptor: | GLYCEROL, IRON/SULFUR CLUSTER, SIROHEME, ... | | Authors: | Schiffer, A, Parey, K, Warkentin, E, Diederichs, K, Huber, H, Stetter, K.O, Kroneck, P.M.H, Ermler, U. | | Deposit date: | 2010-04-19 | | Release date: | 2010-05-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Structure of the dissimilatory sulfite reductase from the hyperthermophilic archaeon Archaeoglobus fulgidus.
J.Mol.Biol., 379, 2008
|
|
4CQL
 
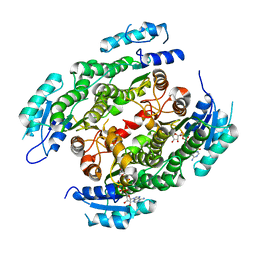 | | Crystal structure of heterotetrameric human ketoacyl reductase complexed with NAD | | Descriptor: | CARBONYL REDUCTASE FAMILY MEMBER 4, ESTRADIOL 17-BETA-DEHYDROGENASE 8, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Venkatesan, R, Sah-Teli, S.K, Awoniyi, L.O, Jiang, G, Prus, P, Kastaniotis, A.J, Hiltunen, J.K, Wierenga, R.K, Chen, Z. | | Deposit date: | 2014-02-19 | | Release date: | 2014-09-10 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Insights Into Mitochondrial Fatty Acid Synthesis from the Structure of Heterotetrameric 3-Ketoacyl-Acp Reductase/3R-Hydroxyacyl-Coa Dehydrogenase.
Nat.Commun., 5, 2014
|
|
4D2H
 
 | |
5TCS
 
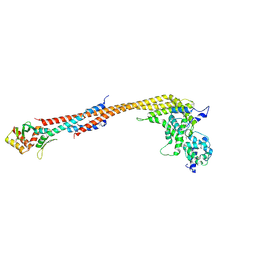 | | Crystal structure of a Dwarf Ndc80 Tetramer | | Descriptor: | Kinetochore protein NDC80, Kinetochore protein NUF2, Kinetochore protein SPC24, ... | | Authors: | Valverde, R, Harrison, S.C. | | Deposit date: | 2016-09-15 | | Release date: | 2016-11-23 | | Last modified: | 2019-11-20 | | Method: | X-RAY DIFFRACTION (2.8313 Å) | | Cite: | Conserved Tetramer Junction in the Kinetochore Ndc80 Complex.
Cell Rep, 17, 2016
|
|
4PXU
 
 | | Structural basis for the assembly of the mitotic motor kinesin-5 into bipolar tetramers | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, Bipolar kinesin KRP-130 | | Authors: | Scholey, J.E, Nithianantham, S, Scholey, J.M, Al-Bassam, J. | | Deposit date: | 2014-03-25 | | Release date: | 2014-04-23 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.601 Å) | | Cite: | Structural basis for the assembly of the mitotic motor Kinesin-5 into bipolar tetramers.
Elife, 3, 2014
|
|
4PXT
 
 | |
4MPW
 
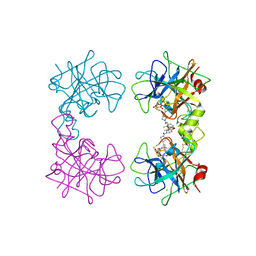 | | Human beta-tryptase co-crystal structure with [(1,1,3,3-tetramethyldisiloxane-1,3-diyl)di-1-benzofuran-3,5-diyl]bis({4-[3-(aminomethyl)phenyl]piperidin-1-yl}methanone) | | Descriptor: | ACETATE ION, CHLORIDE ION, TRIETHYLENE GLYCOL, ... | | Authors: | White, A, Stein, A.J, Suto, R. | | Deposit date: | 2013-09-13 | | Release date: | 2015-03-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Novel, Nonpeptidic, Orally Active Bivalent Inhibitor of Human beta-Tryptase.
Pharmacology, 102, 2018
|
|
4CQM
 
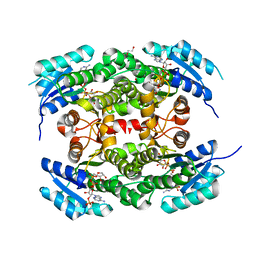 | | Crystal structure of heterotetrameric human ketoacyl reductase complexed with NAD and NADP | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CARBONYL REDUCTASE FAMILY MEMBER 4, ... | | Authors: | Venkatesan, R, SahTeli, S.K, Awoniyi, L.O, Jiang, G, Prus, P, Kastoniotis, A.J, Hiltunen, J.K, Wierenga, R.K, Chen, Z. | | Deposit date: | 2014-02-19 | | Release date: | 2014-09-10 | | Last modified: | 2014-09-17 | | Method: | X-RAY DIFFRACTION (2.339 Å) | | Cite: | Insights Into Mitochondrial Fatty Acid Synthesis from the Structure of Heterotetrameric 3-Ketoacyl-Acp Reductase/3R-Hydroxyacyl-Coa Dehydrogenase.
Nat.Commun., 5, 2014
|
|
3EKZ
 
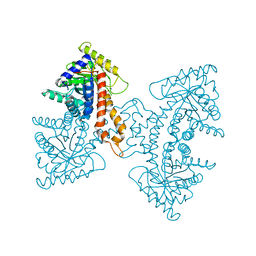 | | Structural Characterization of tetrameric Mycobacterium tuberculosis fructose 1,6-bisphosphate aldolase - substrate binding and catalysis mechanism of a class IIa bacterial aldolase | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, Fructose-bisphosphate aldolase, SN-GLYCEROL-3-PHOSPHATE, ... | | Authors: | Pegan, S, Rukseree, K, Franzblau, S.G, Mesecar, A.D. | | Deposit date: | 2008-09-19 | | Release date: | 2009-02-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Structural basis for catalysis of a tetrameric class IIa fructose 1,6-bisphosphate aldolase from Mycobacterium tuberculosis
J.Mol.Biol., 386, 2009
|
|
1X0L
 
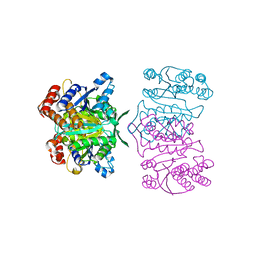 | | Crystal structure of tetrameric homoisocitrate dehydrogenase from an extreme thermophile, Thermus thermophilus | | Descriptor: | Homoisocitrate dehydrogenase | | Authors: | Miyazaki, J, Asada, K, Fushinobu, S, Kuzuyama, T, Nishiyama, M. | | Deposit date: | 2005-03-24 | | Release date: | 2005-10-04 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal Structure of Tetrameric Homoisocitrate Dehydrogenase from an Extreme Thermophile, Thermus thermophilus: Involvement of Hydrophobic Dimer-Dimer Interaction in Extremely High Thermotolerance
J.Bacteriol., 187, 2005
|
|
3ELF
 
 | | Structural Characterization of tetrameric Mycobacterium tuberculosis fructose 1,6-bisphosphate aldolase - substrate binding and catalysis mechanism of a class IIa bacterial aldolase | | Descriptor: | 1,6-FRUCTOSE DIPHOSPHATE (LINEAR FORM), Fructose-bisphosphate aldolase, SODIUM ION, ... | | Authors: | Pegan, S, Rukseree, K, Franzblau, S.G, Mesecar, A.D. | | Deposit date: | 2008-09-22 | | Release date: | 2009-02-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Structural basis for catalysis of a tetrameric class IIa fructose 1,6-bisphosphate aldolase from Mycobacterium tuberculosis
J.Mol.Biol., 386, 2009
|
|
3EKL
 
 | | Structural Characterization of tetrameric Mycobacterium tuberculosis fructose 1,6-bisphosphate aldolase - substrate binding and catalysis mechanism of a class IIa bacterial aldolase | | Descriptor: | 1,3-DIHYDROXYACETONEPHOSPHATE, Fructose-bisphosphate aldolase, SODIUM ION, ... | | Authors: | Pegan, S, Rukseree, K, Franzblau, S.G, Mesecar, A.D. | | Deposit date: | 2008-09-19 | | Release date: | 2009-02-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Structural basis for catalysis of a tetrameric class IIa fructose 1,6-bisphosphate aldolase from Mycobacterium tuberculosis
J.Mol.Biol., 386, 2009
|
|
3EXL
 
 | | Crystal Structure of a p53 Core Tetramer Bound to DNA | | Descriptor: | 5'-D(*DGP*DAP*DGP*DCP*DAP*DTP*DGP*DCP*DTP*DCP*DA)-3', 5'-D(*DTP*DTP*DGP*DAP*DGP*DCP*DAP*DTP*DGP*DCP*DTP*DC)-3', CITRATE ANION, ... | | Authors: | Malecka, K.A. | | Deposit date: | 2008-10-16 | | Release date: | 2008-12-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of a p53 core tetramer bound to DNA.
Oncogene, 28, 2009
|
|
1PES
 
 | | NMR SOLUTION STRUCTURE OF THE TETRAMERIC MINIMUM TRANSFORMING DOMAIN OF P53 | | Descriptor: | TUMOR SUPPRESSOR P53 | | Authors: | Lee, W, Harvey, T.S, Yin, Y, Yau, P, Litchfield, D, Arrowsmith, C.H. | | Deposit date: | 1994-11-24 | | Release date: | 1995-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the tetrameric minimum transforming domain of p53.
Nat.Struct.Biol., 1, 1994
|
|
5VH0
 
 | | RHCC in complex with pyrene | | Descriptor: | 1,2-ETHANEDIOL, Tetrabrachion, pyrene | | Authors: | McDougall, M, Francisco, O, Stetefeld, J. | | Deposit date: | 2017-04-12 | | Release date: | 2018-04-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.055 Å) | | Cite: | Proteinaceous Nano container Encapsulate Polycyclic Aromatic Hydrocarbons.
Sci Rep, 9, 2019
|
|
5VKF
 
 | | RHCC in complex with Naphthalene | | Descriptor: | NAPHTHALENE, SULFATE ION, Tetrabrachion | | Authors: | McDougall, M, Stetefeld, J. | | Deposit date: | 2017-04-21 | | Release date: | 2018-04-25 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.752 Å) | | Cite: | Proteinaceous Nano container Encapsulate Polycyclic Aromatic Hydrocarbons.
Sci Rep, 9, 2019
|
|
3EXJ
 
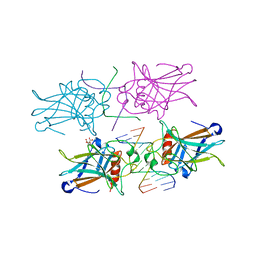 | | Crystal Structure of a p53 Core Tetramer Bound to DNA | | Descriptor: | 5'-D(*DTP*DTP*DGP*DAP*DGP*DCP*DAP*DTP*DGP*DCP*DTP*DC)-3', 5'-D(P*DGP*DAP*DGP*DCP*DAP*DTP*DGP*DCP*DTP*DCP*DA)-3', CITRATE ANION, ... | | Authors: | Malecka, K.A. | | Deposit date: | 2008-10-16 | | Release date: | 2008-12-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a p53 core tetramer bound to DNA.
Oncogene, 28, 2009
|
|
5DOL
 
 | | Crystal structure of YabA amino-terminal domain from Bacillus subtilis | | Descriptor: | Initiation-control protein YabA | | Authors: | Cherrier, M.V, Bazin, A, Jameson, K.H, Wilkinson, A.J, Noirot-Gros, M.F, Terradot, L. | | Deposit date: | 2015-09-11 | | Release date: | 2016-01-20 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Tetramerization and interdomain flexibility of the replication initiation controller YabA enables simultaneous binding to multiple partners.
Nucleic Acids Res., 44, 2016
|
|
1UMR
 
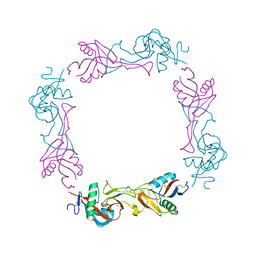 | | Crystal structure of the platelet activator convulxin, a disulfide linked a4b4 cyclic tetramer from the venom of Crotalus durissus terrificus | | Descriptor: | CONVULXIN ALPHA, CONVULXIN BETA | | Authors: | Murakami, M.T, Zela, S.P, Gava, L.M, Michelan-Duarte, S, Cintra, A.C.O, Arni, R.K. | | Deposit date: | 2003-08-28 | | Release date: | 2003-11-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of the Platelet Activator Convulxin, a Disulfide Linked A4B4 Cyclic Tetramer from the Venom of Crotalus Durissus Terrificus
Biochem.Biophys.Res.Commun., 310, 2003
|
|
2EZV
 
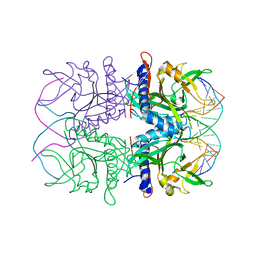 | | Crystal structure of tetrameric restriction endonuclease SfiI bound to cognate DNA. | | Descriptor: | 5'-D(*AP*GP*GP*CP*CP*TP*TP*GP*TP*TP*GP*GP*CP*CP*A)-3', 5'-D(*TP*GP*GP*CP*CP*AP*AP*CP*AP*AP*GP*GP*CP*CP*T)-3', CALCIUM ION, ... | | Authors: | Aggarwal, A.K, Vanamee, E.S, Viadiu, H. | | Deposit date: | 2005-11-10 | | Release date: | 2006-01-24 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | A view of consecutive binding events from structures of tetrameric endonuclease SfiI bound to DNA
Embo J., 24, 2005
|
|
2F03
 
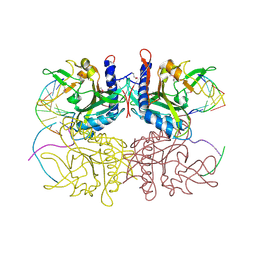 | | Crystal structure of tetrameric restriction endonuclease SfiI in complex with cognate DNA (partial bound form) | | Descriptor: | CALCIUM ION, DNA (5'-D(*AP*AP*TP*AP*GP*GP*CP*CP*TP*TP*GP*TP*TP*GP*GP*CP*CP*AP*CP*AP*T)-3'), DNA (5'-D(*AP*TP*G*TP*GP*GP*CP*CP*AP*AP*CP*AP*AP*GP*GP*CP*CP*TP*AP*TP*T)-3'), ... | | Authors: | Aggarwal, A.K, Vanamee, E.S, Viadiu, H | | Deposit date: | 2005-11-11 | | Release date: | 2007-01-16 | | Last modified: | 2015-09-09 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | A view of consecutive binding events from structures of tetrameric endonuclease SfiI bound to DNA.
Embo J., 24, 2005
|
|
4LCQ
 
 | | The crystal structure of di-Zn dihydropyrimidinase in complex with NCBI | | Descriptor: | (2S)-3-(carbamoylamino)-2-methylpropanoic acid, ZINC ION, dihydropyrimidinase | | Authors: | Hsieh, Y.C, Chen, M.C, Hsu, C.C, Chan, S.I, Yang, Y.S, Chen, C.J. | | Deposit date: | 2013-06-22 | | Release date: | 2013-09-18 | | Last modified: | 2014-02-12 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Crystal structures of vertebrate dihydropyrimidinase and complexes from Tetraodon nigroviridis with lysine carbamylation: metal and structural requirements for post-translational modification and function.
J.Biol.Chem., 288, 2013
|
|
4LCS
 
 | | The crystal structure of di-Zn dihydropyrimidinase in complex with hydantoin | | Descriptor: | Chromosome 8 SCAF14545, whole genome shotgun sequence, ZINC ION, ... | | Authors: | Hsieh, Y.C, Chen, M.C, Hsu, C.C, Chan, S.I, Yang, Y.S. | | Deposit date: | 2013-06-23 | | Release date: | 2013-09-18 | | Last modified: | 2014-02-12 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structures of vertebrate dihydropyrimidinase and complexes from Tetraodon nigroviridis with lysine carbamylation: metal and structural requirements for post-translational modification and function.
J.Biol.Chem., 288, 2013
|
|
