6WA2
 
 | | Crystal structure of EGFR(T790M/V948R) in complex with LN3753 | | Descriptor: | CHLORIDE ION, Epidermal growth factor receptor, N-(3-{5-[2-(acetylamino)pyridin-4-yl]-2-(methylsulfanyl)-1H-imidazol-4-yl}phenyl)-2-fluoro-5-hydroxybenzamide | | Authors: | Heppner, D.E, Eck, M.J. | | Deposit date: | 2020-03-24 | | Release date: | 2021-03-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design of a "Two-in-One" Mutant-Selective Epidermal Growth Factor Receptor Inhibitor That Spans the Orthosteric and Allosteric Sites.
J.Med.Chem., 65, 2022
|
|
6WAK
 
 | | A crystal structure of EGFR(T790M/V948R) in complex with LN3754 | | Descriptor: | Epidermal growth factor receptor, MAGNESIUM ION, N-(3-{5-[2-(acetylamino)pyridin-4-yl]-2-(methylsulfanyl)-1H-imidazol-4-yl}phenyl)-2-[(1-oxo-1,3-dihydro-2H-isoindol-2-yl)methyl]benzamide, ... | | Authors: | Heppner, D.E, Eck, M.J. | | Deposit date: | 2020-03-25 | | Release date: | 2021-03-31 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design of a "Two-in-One" Mutant-Selective Epidermal Growth Factor Receptor Inhibitor That Spans the Orthosteric and Allosteric Sites.
J.Med.Chem., 65, 2022
|
|
1PH0
 
 | | Non-carboxylic Acid-Containing Inhibitor of PTP1B Targeting the Second Phosphotyrosine Site | | Descriptor: | 2-{4-[2-(S)-ALLYLOXYCARBONYLAMINO-3-{4-[(2-CARBOXY-PHENYL)-OXALYL-AMINO]-PHENYL}-PROPIONYLAMINO]-BUTOXY}-6-HYDROXY-BENZ OIC ACID METHYL ESTER, Protein-tyrosine phosphatase, non-receptor type 1 | | Authors: | Liu, G, Xin, Z, Liang, H, Abad-Zapatero, C, Hajduk, P, Janowick, D, Szczepankiewicz, B, Pei, Z, Hutchins, C.W, Ballaron, S.J. | | Deposit date: | 2003-05-29 | | Release date: | 2003-07-29 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Selective Protein Tyrosine Phosphatase 1B Inhibitors: Targeting the Second Phosphotyrosine Binding Site with Non-Carboxylic Acid-Containing Ligands.
J.Med.Chem., 46, 2003
|
|
6XZ5
 
 | |
2IHR
 
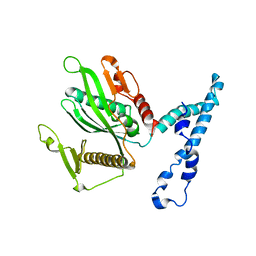 | | RF2 of Thermus thermophilus | | Descriptor: | Peptide chain release factor 2 | | Authors: | Dobbek, H, Voertler, C.S, Sprinzl, M. | | Deposit date: | 2006-09-27 | | Release date: | 2007-02-20 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Release factors 2 from Escherichia coli and Thermus thermophilus: structural, spectroscopic and microcalorimetric studies.
Nucleic Acids Res., 35, 2007
|
|
6XVR
 
 | | Human myelin protein P2 mutant L35S | | Descriptor: | Myelin P2 protein, PALMITIC ACID, SULFATE ION | | Authors: | Ruskamo, S, Lehtimaki, M, Kursula, P. | | Deposit date: | 2020-01-22 | | Release date: | 2020-04-08 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Cryo-EM, X-ray diffraction, and atomistic simulations reveal determinants for the formation of a supramolecular myelin-like proteolipid lattice.
J.Biol.Chem., 295, 2020
|
|
7ODW
 
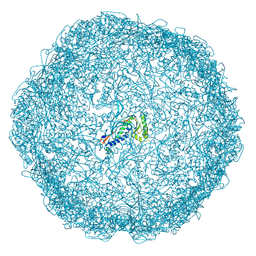 | |
6ZIO
 
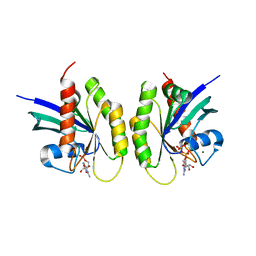 | |
7OG5
 
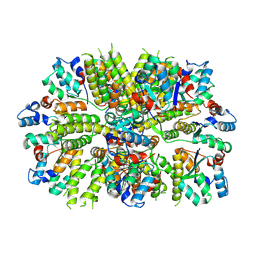 | |
6Y6X
 
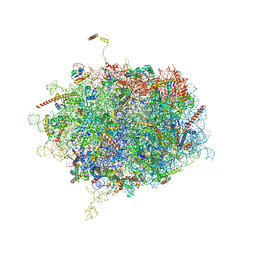 | | Tetracenomycin X bound to the human ribosome | | Descriptor: | 28S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Buschauer, R, Cheng, J, Berninghausen, O, Beckmann, R, Wilson, D.N. | | Deposit date: | 2020-02-27 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Tetracenomycin X inhibits translation by binding within the ribosomal exit tunnel.
Nat.Chem.Biol., 16, 2020
|
|
7OGG
 
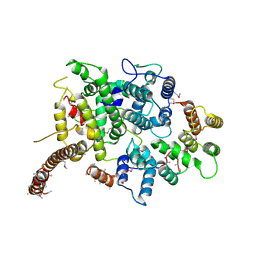 | | Nse5/6 complex | | Descriptor: | DNA repair protein KRE29,DNA repair protein KRE29,DNA repair protein KRE29, Non-structural maintenance of chromosome element 5,Non-structural maintenance of chromosome element 5,Non-structural maintenance of chromosome element 5 | | Authors: | Basquin, J, Taschner, M, Gruber, S. | | Deposit date: | 2021-05-06 | | Release date: | 2021-07-07 | | Last modified: | 2021-08-11 | | Method: | X-RAY DIFFRACTION (3.29 Å) | | Cite: | Nse5/6 inhibits the Smc5/6 ATPase and modulates DNA substrate binding.
Embo J., 40, 2021
|
|
1OFE
 
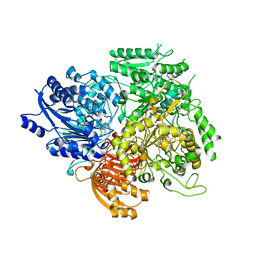 | |
6YUM
 
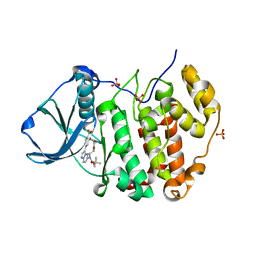 | | CK2 alpha bound to unclosed Macrocycle | | Descriptor: | 4-[5-[2-(2-hydroxyethyloxy)ethyl-[(2-methylpropan-2-yl)oxycarbonyl]amino]pyrazolo[1,5-a]pyrimidin-3-yl]-2-oxidanyl-benzoic acid, Casein kinase II subunit alpha, SULFATE ION | | Authors: | Kraemer, A, Hanke, T, Kurz, C, Celik, I, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-04-27 | | Release date: | 2020-07-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Optimization of pyrazolo[1,5-a]pyrimidines lead to the identification of a highly selective casein kinase 2 inhibitor.
Eur.J.Med.Chem., 208, 2020
|
|
2IGM
 
 | | Crystal structure of recombinant pyranose 2-oxidase H548N mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, Pyranose oxidase | | Authors: | Divne, C. | | Deposit date: | 2006-09-22 | | Release date: | 2006-10-10 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for substrate binding and regioselective oxidation of monosaccharides at c3 by pyranose 2-oxidase.
J.Biol.Chem., 281, 2006
|
|
6Z30
 
 | | Human cation-independent mannose 6-phosphate/ IGF2 receptor domains 9-10 | | Descriptor: | Cation-independent mannose-6-phosphate receptor, alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Bochel, A.J, Williams, C, Crump, M.P. | | Deposit date: | 2020-05-19 | | Release date: | 2020-08-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure of the Human Cation-Independent Mannose 6-Phosphate/IGF2 Receptor Domains 7-11 Uncovers the Mannose 6-Phosphate Binding Site of Domain 9.
Structure, 28, 2020
|
|
1M1M
 
 | |
6Z9J
 
 | |
7O70
 
 | | KRasG12C ligand complex | | Descriptor: | 1-[(4R,7S)-12-chloro-14-fluoro-13-(2-fluoro-6-hydroxyphenyl)-4-methyl-10-oxa-2,5,16,18-tetrazatetracyclo[9.7.1.0^(2,7).0^(15,19)]nonadeca-1(18),11,13,15(19),16-pentaen-5-en-1-one-yl]prop-2, CALCIUM ION, GTPase KRas, ... | | Authors: | Phillips, C. | | Deposit date: | 2021-04-12 | | Release date: | 2022-04-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.18 Å) | | Cite: | Discovery of AZD4625, a Covalent Allosteric Inhibitor of the Mutant GTPase KRAS G12C .
J.Med.Chem., 65, 2022
|
|
7O83
 
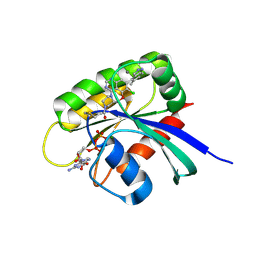 | | KRasG12C ligand complex | | Descriptor: | 1-[(7S)-11-chloro-12-(5-methyl-1H-indazol-4-yl)-9-oxa-2,5,15,17-tetrazatetracyclo[8.7.1.02,7.014,18]octadeca-1(17),10,12,14(18),15-pentaen-5-yl]prop-2-en-1-one, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Phillips, C, Breed, J. | | Deposit date: | 2021-04-14 | | Release date: | 2022-04-20 | | Last modified: | 2022-05-25 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Discovery of AZD4625, a Covalent Allosteric Inhibitor of the Mutant GTPase KRAS G12C .
J.Med.Chem., 65, 2022
|
|
7OAE
 
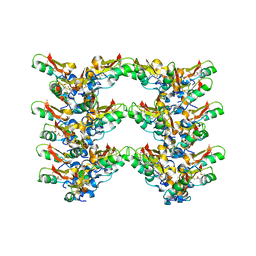 | | Cryo-EM structure of the plectasin fibril (double strands) | | Descriptor: | Fungal defensin plectasin | | Authors: | Effantin, G. | | Deposit date: | 2021-04-19 | | Release date: | 2022-04-27 | | Last modified: | 2022-11-16 | | Method: | ELECTRON MICROSCOPY (2 Å) | | Cite: | pH- and concentration-dependent supramolecular assembly of a fungal defensin plectasin variant into helical non-amyloid fibrils.
Nat Commun, 13, 2022
|
|
2IGN
 
 | | Crystal structure of recombinant pyranose 2-oxidase H167A mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, FLAVIN-ADENINE DINUCLEOTIDE, Pyranose oxidase | | Authors: | Divne, C. | | Deposit date: | 2006-09-22 | | Release date: | 2006-10-10 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural basis for substrate binding and regioselective oxidation of monosaccharides at c3 by pyranose 2-oxidase.
J.Biol.Chem., 281, 2006
|
|
7OAG
 
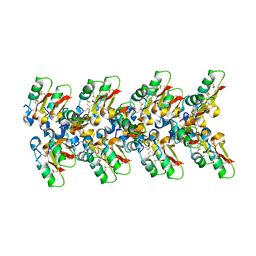 | | Cryo-EM structure of the plectasin fibril (single strand) | | Descriptor: | Fungal defensin plectasin | | Authors: | Effantin, G. | | Deposit date: | 2021-04-19 | | Release date: | 2022-04-27 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | pH- and concentration-dependent supramolecular assembly of a fungal defensin plectasin variant into helical non-amyloid fibrils.
Nat Commun, 13, 2022
|
|
7NQD
 
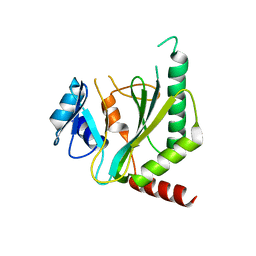 | |
7NQE
 
 | |
7NQF
 
 | |
