2JJ9
 
 | | Crystal structure of myosin-2 in complex with ADP-metavanadate | | Descriptor: | ADP METAVANADATE, MAGNESIUM ION, MYOSIN-2 HEAVY CHAIN | | Authors: | Fedorov, R, Boehl, M, Tsiavaliaris, G, Hartmann, F.K, Baruch, P, Brenner, B, Martin, R, Knoelker, H.J, Gutzeit, H.O, Manstein, D.J. | | Deposit date: | 2008-03-25 | | Release date: | 2009-01-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Mechanism of Pentabromopseudilin Inhibition of Myosin Motor Activity.
Nat.Struct.Mol.Biol., 16, 2009
|
|
2KIA
 
 | | Solution structure of Myosin VI C-terminal cargo-binding domain | | Descriptor: | Myosin-VI | | Authors: | Feng, W, Yu, C, Wei, Z, Miyanoiri, Y, Zhang, M. | | Deposit date: | 2009-04-29 | | Release date: | 2009-09-29 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Myosin VI undergoes cargo-mediated dimerization
Cell(Cambridge,Mass.), 138, 2009
|
|
1TTX
 
 | | Solution Structure of human beta parvalbumin (oncomodulin) refined with a paramagnetism based strategy | | Descriptor: | CALCIUM ION, Oncomodulin | | Authors: | Babini, E, Bertini, I, Capozzi, F, Del Bianco, C, Hollender, D, Kiss, T, Luchinat, C, Quattrone, A, Structural Proteomics in Europe (SPINE) | | Deposit date: | 2004-06-23 | | Release date: | 2005-01-18 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of Human beta-Parvalbumin and Structural Comparison with Its Paralog alpha-Parvalbumin and with Their Rat Orthologs(,)
Biochemistry, 43, 2004
|
|
1C7W
 
 | | NMR SOLUTION STRUCTURE OF THE CALCIUM-BOUND C-TERMINAL DOMAIN (W81-S161) OF CALCIUM VECTOR PROTEIN FROM AMPHIOXUS | | Descriptor: | CALCIUM VECTOR PROTEIN | | Authors: | Theret, I, Baladi, S, Cox, J.A, Sakamoto, H, Craescu, C.T. | | Deposit date: | 2000-03-27 | | Release date: | 2000-04-12 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Sequential calcium binding to the regulatory domain of calcium vector protein reveals functional asymmetry and a novel mode of structural rearrangement.
Biochemistry, 39, 2000
|
|
1C7V
 
 | | NMR SOLUTION STRUCTURE OF THE CALCIUM-BOUND C-TERMINAL DOMAIN (W81-S161) OF CALCIUM VECTOR PROTEIN FROM AMPHIOXUS | | Descriptor: | CALCIUM VECTOR PROTEIN | | Authors: | Theret, I, Baladi, S, Cox, J.A, Sakamoto, H, Craescu, C.T. | | Deposit date: | 2000-03-27 | | Release date: | 2000-04-12 | | Last modified: | 2023-12-27 | | Method: | SOLUTION NMR | | Cite: | Sequential calcium binding to the regulatory domain of calcium vector protein reveals functional asymmetry and a novel mode of structural rearrangement.
Biochemistry, 39, 2000
|
|
2SCP
 
 | |
2SAS
 
 | |
2PHK
 
 | | THE CRYSTAL STRUCTURE OF A PHOSPHORYLASE KINASE PEPTIDE SUBSTRATE COMPLEX: KINASE SUBSTRATE RECOGNITION | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GLYCEROL, MANGANESE (II) ION, ... | | Authors: | Lowe, E.D, Noble, M.E.M, Skamnaki, V.T, Oikonomakos, N.G, Owen, D.J, Johnson, L.N. | | Deposit date: | 1998-06-18 | | Release date: | 1999-01-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The crystal structure of a phosphorylase kinase peptide substrate complex: kinase substrate recognition.
EMBO J., 16, 1997
|
|
1E8A
 
 | | The three-dimensional structure of human S100A12 | | Descriptor: | CALCIUM ION, S100A12 | | Authors: | Moroz, O.V, Antson, A.A, Murshudov, G.N, Maitland, N.J, Dodson, G.G, Wilson, K.S, Skibshoj, I, Lukanidin, E.M, Bronstein, I.B. | | Deposit date: | 2000-09-18 | | Release date: | 2001-01-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Three-Dimensional Structure of Human S100A12
Acta Crystallogr.,Sect.D, 57, 2001
|
|
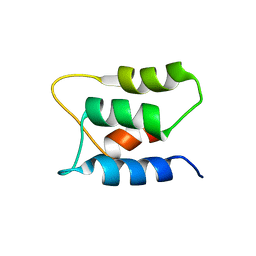
1B8C
 
 | | PARVALBUMIN | | Descriptor: | MAGNESIUM ION, PROTEIN (PARVALBUMIN) | | Authors: | Cates, M.S, Berry, M.B, Ho, E.L, Li, Q, Potter, J.D, Phillips Jr, G.N. | | Deposit date: | 1999-01-29 | | Release date: | 1999-02-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Metal-ion affinity and specificity in EF-hand proteins: coordination geometry and domain plasticity in parvalbumin.
Structure Fold.Des., 7, 1999
|
|
1FHG
 
 | | HIGH RESOLUTION REFINEMENT OF TELOKIN | | Descriptor: | TELOKIN | | Authors: | Tomchick, D.R, Minor, W, Kiyatkin, A, Lewinski, K, Somlyo, A.V, Somlyo, A.P. | | Deposit date: | 2000-08-01 | | Release date: | 2000-08-23 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray structure determination of telokin, the C-terminal domain of myosin light chain kinase, at 2.8 A resolution.
J.Mol.Biol., 227, 1992
|
|
2K7C
 
 | |
2LV7
 
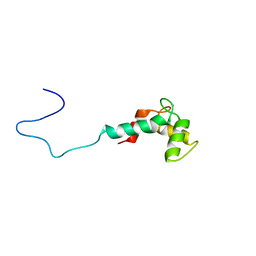 | | Solution structure of Ca2+-bound CaBP7 N-terminal doman | | Descriptor: | CALCIUM ION, Calcium-binding protein 7 | | Authors: | Mccue, H.V, Patel, P, Herbert, A.P, Lian, L, Burgoyne, R.D, Haynes, L.P. | | Deposit date: | 2012-06-29 | | Release date: | 2012-09-26 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of the Ca2+-bound N-terminal Domain of CaBP7: A REGULATOR OF GOLGI TRAFFICKING.
J.Biol.Chem., 287, 2012
|
|
2M7M
 
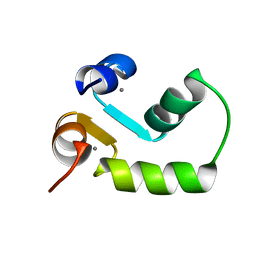 | | N-terminal domain of EhCaBP1 structure | | Descriptor: | CALCIUM ION, Calcium-binding protein | | Authors: | Chary, K.V, Rout, A.K, Patel, S, Bhattacharya, A. | | Deposit date: | 2013-04-26 | | Release date: | 2013-06-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Functional Manipulation of a Calcium-binding Protein from Entamoeba histolytica Guided by Paramagnetic NMR.
J.Biol.Chem., 288, 2013
|
|
2M7N
 
 | | C-terminal structure of (Y81F)-EhCaBP1 | | Descriptor: | CALCIUM ION, Calcium-binding protein | | Authors: | Chary, K.V, Rout, A.K, Patel, S, Bhattacharya, A. | | Deposit date: | 2013-04-26 | | Release date: | 2013-06-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Functional Manipulation of a Calcium-binding Protein from Entamoeba histolytica Guided by Paramagnetic NMR.
J.Biol.Chem., 288, 2013
|
|
2JUL
 
 | | NMR Structure of DREAM | | Descriptor: | CALCIUM ION, Calsenilin | | Authors: | Ames, J. | | Deposit date: | 2007-08-30 | | Release date: | 2008-04-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | NMR structure of DREAM: Implications for Ca(2+)-dependent DNA binding and protein dimerization.
Biochemistry, 47, 2008
|
|
2KFX
 
 | |
2KOG
 
 | | lipid-bound synaptobrevin solution NMR structure | | Descriptor: | Vesicle-associated membrane protein 2 | | Authors: | Ellena, J.F, Liang, B, Wiktor, M, Stein, A, Cafiso, D.S, Jahn, R, Tamm, L.K. | | Deposit date: | 2009-09-22 | | Release date: | 2009-12-01 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Dynamic structure of lipid-bound synaptobrevin suggests a nucleation-propagation mechanism for trans-SNARE complex formation.
Proc.Natl.Acad.Sci.USA, 106, 2009
|
|
2KRD
 
 | | Solution Structure of the Regulatory Domain of Human Cardiac Troponin C in Complex with the Switch Region of cardiac Troponin I and W7 | | Descriptor: | CALCIUM ION, N-(6-AMINOHEXYL)-5-CHLORO-1-NAPHTHALENESULFONAMIDE, Troponin C, ... | | Authors: | Oleszczuk, M, Robertson, I.M, Li, M.X, Sykes, B.D. | | Deposit date: | 2009-12-16 | | Release date: | 2010-02-16 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the regulatory domain of human cardiac troponin C in complex with the switch region of cardiac troponin I and W7: the basis of W7 as an inhibitor of cardiac muscle contraction.
J.MOL.CELL.CARDIOL., 48, 2010
|
|
2NMP
 
 | | Crystal structure of human Cystathionine gamma lyase | | Descriptor: | Cystathionine gamma-lyase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Karlberg, T, Uppenberg, J, Arrowsmith, C, Berglund, H, Busam, R.D, Collins, R, Edwards, A, Ericsson, U.B, Flodin, S, Flores, A, Graslund, S, Hallberg, B.M, Hammarstrom, M, Hogbom, M, Johansson, I, Kotenyova, T, Magnusdottir, A, Moche, M, Nilsson, M.E, Nordlund, P, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Stenmark, P, Sundstrom, M, Thorsell, A.G, van-den-Berg, S, Wallden, K, Weigelt, J, Holmberg-Schiavone, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2006-10-23 | | Release date: | 2006-11-07 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural basis for the inhibition mechanism of human cystathionine gamma-lyase, an enzyme responsible for the production of H(2)S
J.Biol.Chem., 284, 2009
|
|
2NCP
 
 | |
2NCO
 
 | |
2NA0
 
 | |
2JU0
 
 | | Structure of Yeast Frequenin bound to PdtIns 4-kinase | | Descriptor: | CALCIUM ION, Calcium-binding protein NCS-1, Phosphatidylinositol 4-kinase PIK1 | | Authors: | Ames, J. | | Deposit date: | 2007-08-11 | | Release date: | 2007-08-28 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Structural insights into activation of phosphatidylinositol 4-kinase (Pik1) by yeast frequenin (Frq1).
J.Biol.Chem., 282, 2007
|
|
2L4I
 
 | | The Solution Structure of Magnesium bound CIB1 | | Descriptor: | Calcium and integrin-binding protein 1, MAGNESIUM ION | | Authors: | Huang, H, Vogel, H.J. | | Deposit date: | 2010-10-06 | | Release date: | 2011-03-09 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution Structures of Ca2+-CIB1 and Mg2+-CIB1 and Their Interactions with the Platelet Integrin {alpha}IIb Cytoplasmic Domain.
J.Biol.Chem., 286, 2011
|
|
