1PLS
 
 | | SOLUTION STRUCTURE OF A PLECKSTRIN HOMOLOGY DOMAIN | | Descriptor: | PLECKSTRIN HOMOLOGY DOMAIN | | Authors: | Yoon, H.S, Hajduk, P.J, Petros, A.M, Olejniczak, E.T, Meadows, R.P, Fesik, S.W. | | Deposit date: | 1994-05-03 | | Release date: | 1995-06-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of a pleckstrin-homology domain.
Nature, 369, 1994
|
|
6R6H
 
 | | Structural basis of Cullin-2 RING E3 ligase regulation by the COP9 signalosome | | Descriptor: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | Authors: | Morris, E.P, Faull, S.V, Lau, A.M.C, Politis, A, Beuron, F, Cronin, N. | | Deposit date: | 2019-03-27 | | Release date: | 2019-08-28 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (8.4 Å) | | Cite: | Structural basis of Cullin 2 RING E3 ligase regulation by the COP9 signalosome.
Nat Commun, 10, 2019
|
|
1JLT
 
 | | Vipoxin Complex | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, PHOSPHOLIPASE A2, ... | | Authors: | Banumathi, S, Rajashankar, K.R, Notzel, C, Aleksiev, B, Singh, T.P, Genov, N, Betzel, C. | | Deposit date: | 2001-07-16 | | Release date: | 2001-10-31 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structure of the neurotoxic complex vipoxin at 1.4 A resolution.
Acta Crystallogr.,Sect.D, 57, 2001
|
|
5EXA
 
 | | Small-molecule stabilization of the 14-3-3/Gab2 PPI interface | | Descriptor: | 14-3-3 protein zeta/delta, BENZOIC ACID, Fusicoccin A-THF derivative, ... | | Authors: | Bier, D, Ottmann, C. | | Deposit date: | 2015-11-23 | | Release date: | 2016-05-04 | | Last modified: | 2018-12-19 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Small-Molecule Stabilization of the 14-3-3/Gab2 Protein-Protein Interaction (PPI) Interface.
Chemmedchem, 11, 2016
|
|
1W2N
 
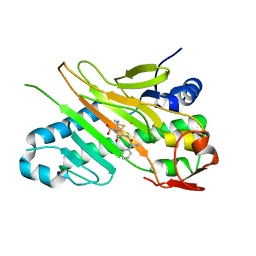 | | Deacetoxycephalosporin C synthase (with a N-terminal his-tag) in complex with Fe(II) and ampicillin | | Descriptor: | (2S,6R)-6-{[(2R)-2-AMINO-2-PHENYLETHANOYL]AMINO}-3,3-DIMETHYL-7-OXO-4-THIA-1-AZABICYCLO[3.2.0]HEPTANE-2-CARBOXYLIC ACID, DEACETOXYCEPHALOSPORIN C SYNTHASE, FE (III) ION | | Authors: | Oster, L.M, Terwisscha Van Scheltinga, A.C, Valegard, K, Mackenzie Hose, A, Dubus, A, Hajdu, J, Andersson, I. | | Deposit date: | 2004-07-07 | | Release date: | 2004-09-30 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Conformational Flexibility of the C Terminus with Implications for Substrate Binding and Catalysis Revealed in a New Crystal Form of Deacetoxycephalosporin C Synthase
J.Mol.Biol., 343, 2004
|
|
5EWZ
 
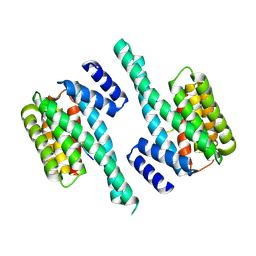 | | Small-molecule stabilization of the 14-3-3/Gab2 PPI interface | | Descriptor: | 14-3-3 protein zeta/delta, BENZOIC ACID, GRB2-associated-binding protein 2 | | Authors: | Bier, D, Ottmann, C. | | Deposit date: | 2015-11-23 | | Release date: | 2016-05-04 | | Last modified: | 2018-12-19 | | Method: | X-RAY DIFFRACTION (2.34 Å) | | Cite: | Small-Molecule Stabilization of the 14-3-3/Gab2 Protein-Protein Interaction (PPI) Interface.
Chemmedchem, 11, 2016
|
|
5ETJ
 
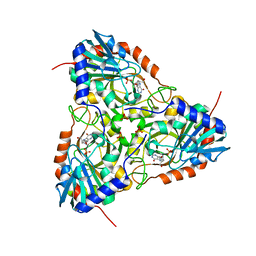 | | Crystal structure of purine nucleoside phosphorylase (E258D, L261A) mutant from human complexed with DADMe-ImmG and phosphate | | Descriptor: | 2-amino-7-{[(3R,4R)-3-hydroxy-4-(hydroxymethyl)pyrrolidin-1-yl]methyl}-3,5-dihydro-4H-pyrrolo[3,2-d]pyrimidin-4-one, PHOSPHATE ION, Purine nucleoside phosphorylase | | Authors: | Cameron, S.A, Suarez, J, Schramm, V.L, Almo, S.C. | | Deposit date: | 2015-11-17 | | Release date: | 2016-09-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Modulating Enzyme Catalysis through Mutations Designed to Alter Rapid Protein Dynamics.
J.Am.Chem.Soc., 138, 2016
|
|
1JAU
 
 | |
1NYY
 
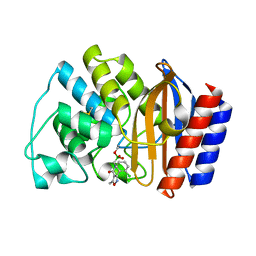 | | Crystal Structure of the complex between M182T mutant of TEM-1 and a boronic acid inhibitor (105) | | Descriptor: | Beta-lactamase TEM, N-[5-METHYL-3-O-TOLYL-ISOXAZOLE-4-CARBOXYLIC ACID AMIDE] BORONIC ACID | | Authors: | Wang, X, Minasov, G, Blazquez, J, Caselli, E, Prati, F, Shoichet, B.K. | | Deposit date: | 2003-02-14 | | Release date: | 2003-08-26 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Recognition and resistance in TEM beta-lactamase
Biochemistry, 42, 2003
|
|
1JJZ
 
 | |
8EVA
 
 | |
8EVB
 
 | |
8EV9
 
 | |
8EV8
 
 | |
8EVC
 
 | |
1JE3
 
 | | Solution Structure of EC005 from Escherichia coli | | Descriptor: | HYPOTHETICAL 8.6 KDA PROTEIN IN AMYA-FLIE INTERGENIC REGION | | Authors: | Yee, A, Gutierrez, P, Kozlov, G, Denisov, A, Gehring, K, Arrowsmith, C. | | Deposit date: | 2001-06-15 | | Release date: | 2002-03-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | An NMR approach to structural proteomics.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
3DQN
 
 | |
3EJ0
 
 | |
4KH6
 
 | |
1NZR
 
 | | CRYSTAL STRUCTURE OF THE AZURIN MUTANT NICKEL-TRP48MET FROM PSEUDOMONAS AERUGINOSA AT 2.2 ANGSTROMS RESOLUTION | | Descriptor: | AZURIN, NICKEL (II) ION, NITRATE ION | | Authors: | Tsai, L.-C, Sjolin, L, Langer, V, Bonander, N, Karlsson, B.G, Vanngard, T, Hammann, C, Nar, H. | | Deposit date: | 1994-12-09 | | Release date: | 1995-02-27 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of the azurin mutant nickel-Trp48Met from Pseudomonas aeruginosa at 2.2 A resolution.
Acta Crystallogr.,Sect.D, 51, 1995
|
|
1RG9
 
 | | S-Adenosylmethionine synthetase complexed with SAM and PPNP | | Descriptor: | (DIPHOSPHONO)AMINOPHOSPHONIC ACID, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Komoto, J, Yamada, T, Takata, Y, Markham, G.D, Takusagawa, F. | | Deposit date: | 2003-11-11 | | Release date: | 2004-03-02 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of the S-adenosylmethionine synthetase ternary complex: a novel catalytic mechanism of s-adenosylmethionine synthesis from ATP and MET.
Biochemistry, 43, 2004
|
|
1ZKM
 
 | | Structural Analysis of Escherichia Coli ThiF | | Descriptor: | Adenylyltransferase thiF, ZINC ION | | Authors: | Duda, D.M, Walden, H, Sfondouris, J, Schulman, B.A. | | Deposit date: | 2005-05-03 | | Release date: | 2005-06-14 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Structural Analysis of Escherichia Coli ThiF.
J.Mol.Biol., 349, 2005
|
|
3CJB
 
 | | Actin dimer cross-linked by V. cholerae MARTX toxin and complexed with Gelsolin-segment 1 | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin, alpha skeletal muscle, ... | | Authors: | Sawaya, M.R, Kudryashov, D.S, Pashkov, I, Reisler, E, Yeates, T.O. | | Deposit date: | 2008-03-12 | | Release date: | 2008-03-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.21 Å) | | Cite: | Connecting actin monomers by iso-peptide bond is a toxicity mechanism of the Vibrio cholerae MARTX toxin.
Proc.Natl.Acad.Sci.USA, 105, 2008
|
|
5KMG
 
 | | Near-atomic cryo-EM structure of PRC1 bound to the microtubule | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Kellogg, E.H, Howes, S, Ti, S.-C, Ramirez-Aportela, E, Kapoor, T.M, Chacon, P, Nogales, E. | | Deposit date: | 2016-06-27 | | Release date: | 2016-08-03 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Near-atomic cryo-EM structure of PRC1 bound to the microtubule.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
6SGB
 
 | | mt-SSU assemblosome of Trypanosoma brucei | | Descriptor: | 9S rRNA, GUANOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Saurer, M, Ramrath, D.J.F, Niemann, M, Calderaro, S, Prange, C, Mattei, S, Scaiola, A, Leitner, A, Bieri, P, Horn, E.K, Leibundgut, M, Boehringer, D, Schneider, A, Ban, N. | | Deposit date: | 2019-08-03 | | Release date: | 2019-09-25 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mitoribosomal small subunit biogenesis in trypanosomes involves an extensive assembly machinery.
Science, 365, 2019
|
|
