6V37
 
 | |
8PNW
 
 | | Crystal structure of D-amino acid aminotransferase from Blastococcus saxobsidens in holo form with PLP | | Descriptor: | Branched-chain amino acid aminotransferase/4-amino-4-deoxychorismate lyase, CHLORIDE ION, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Matyuta, I.O, Boyko, K.M, Nikolaeva, A.Y, Shilova, S.A, Popov, V.O. | | Deposit date: | 2023-07-03 | | Release date: | 2023-10-25 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Expanded Substrate Specificity in D-Amino Acid Transaminases: A Case Study of Transaminase from Blastococcus saxobsidens.
Int J Mol Sci, 24, 2023
|
|
8CZK
 
 | |
6UTA
 
 | | Crystal structure of Z004 iGL Fab in complex with ZIKV EDIII | | Descriptor: | Env, Z004 iGL Fab heavy chain, Z004 iGL Fab light chain | | Authors: | Esswein, S.R, Gristick, H.B, Keeffe, J.R, Bjorkman, P.J. | | Deposit date: | 2019-10-29 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structural basis for Zika envelope domain III recognition by a germline version of a recurrent neutralizing antibody.
Proc.Natl.Acad.Sci.USA, 117, 2020
|
|
7T1I
 
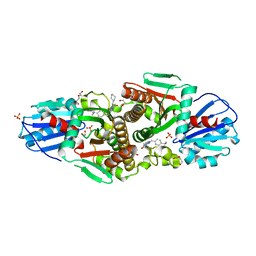 | | Crystal structure of CAB1 Pantothenate Kinase from Saccharomyces cerevisiae in complex with compound YU385597 | | Descriptor: | (8S)-2-{[(4-tert-butylphenyl)methyl]amino}-5-[(piperidin-1-yl)methyl][1,2,4]triazolo[1,5-a]pyrimidin-7(6H)-one, 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Gihaz, S, Ben Mamoun, C. | | Deposit date: | 2021-12-02 | | Release date: | 2022-10-05 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | High-resolution crystal structure and chemical screening reveal pantothenate kinase as a new target for antifungal development.
Structure, 30, 2022
|
|
7WRK
 
 | |
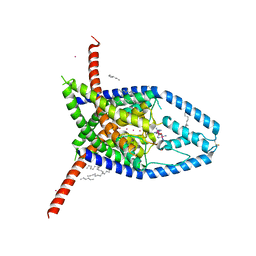
6V3I
 
 | |
6PN0
 
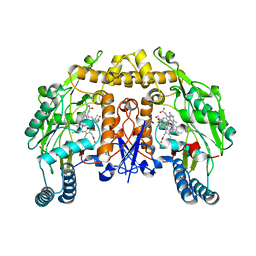 | | Structure of rat neuronal nitric oxide synthase heme domain in complex with 7-(3-(Aminomethyl)-4-ethoxyphenyl)-4-methylquinolin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 7-[3-(aminomethyl)-4-ethoxyphenyl]-4-methylquinolin-2-amine, ACETATE ION, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2019-07-02 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.229 Å) | | Cite: | First Contact: 7-Phenyl-2-Aminoquinolines, Potent and Selective Neuronal Nitric Oxide Synthase Inhibitors That Target an Isoform-Specific Aspartate.
J.Med.Chem., 63, 2020
|
|
8DJX
 
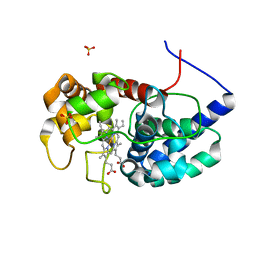 | | Cytosolic ascorbate peroxidase from Sorghum bicolor - Compound II | | Descriptor: | L-ascorbate peroxidase, PROTOPORPHYRIN IX CONTAINING FE, SODIUM ION, ... | | Authors: | Zhang, B, Kang, C. | | Deposit date: | 2022-07-01 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | A sorghum ascorbate peroxidase with four binding sites has activity against ascorbate and phenylpropanoids.
Plant Physiol., 192, 2023
|
|
7O0C
 
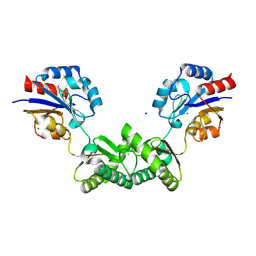 | | Human phosphomannomutase 2 (PMM2) wild-type in apo state | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Phosphomannomutase 2, ... | | Authors: | Ramon-Maiques, S, Briso-Montiano, A, Del Cano-Ochoa, F, Vilas, A, Perez, B, Rubio, V. | | Deposit date: | 2021-03-26 | | Release date: | 2022-02-02 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Insight on molecular pathogenesis and pharmacochaperoning potential in phosphomannomutase 2 deficiency, provided by novel human phosphomannomutase 2 structures.
J Inherit Metab Dis, 45, 2022
|
|
7W2C
 
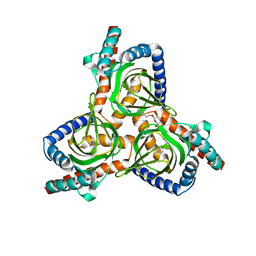 | | The closed conformation of the sigma-1 receptor from Xenopus laevis complexed with PRE084 | | Descriptor: | 2-morpholin-4-ylethyl 1-phenylcyclohexane-1-carboxylate, GOLD ION, Sigma non-opioid intracellular receptor 1, ... | | Authors: | Meng, F, Sun, Z, Zhou, X. | | Deposit date: | 2021-11-23 | | Release date: | 2022-03-16 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.335 Å) | | Cite: | An open-like conformation of the sigma-1 receptor reveals its ligand entry pathway.
Nat Commun, 13, 2022
|
|
6PNA
 
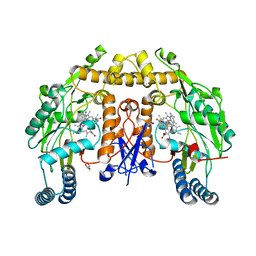 | | Structure of human neuronal nitric oxide synthase R354A/G357D mutant heme domain in complex with 7-(4-(2-Aminoethyl)phenyl)-4-methylquinolin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 7-[4-(2-aminoethyl)phenyl]-4-methylquinolin-2-amine, GLYCEROL, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2019-07-02 | | Release date: | 2020-04-29 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | First Contact: 7-Phenyl-2-Aminoquinolines, Potent and Selective Neuronal Nitric Oxide Synthase Inhibitors That Target an Isoform-Specific Aspartate.
J.Med.Chem., 63, 2020
|
|
6AUZ
 
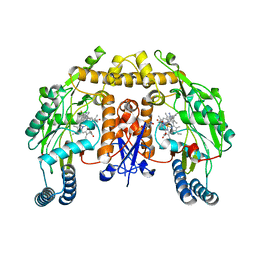 | | Structure of human neuronal nitric oxide synthase R354A/G357D mutant heme domain in complex with 6-(3-Fluoro-5-(3-(methylamino)propyl)phenethyl)-4-methylpyridin-2-amine | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, 6-(2-{3-fluoro-5-[3-(methylamino)propyl]phenyl}ethyl)-4-methylpyridin-2-amine, Nitric oxide synthase, ... | | Authors: | Li, H, Poulos, T.L. | | Deposit date: | 2017-09-01 | | Release date: | 2018-07-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Improvement of Cell Permeability of Human Neuronal Nitric Oxide Synthase Inhibitors Using Potent and Selective 2-Aminopyridine-Based Scaffolds with a Fluorobenzene Linker.
J. Med. Chem., 60, 2017
|
|
7T5V
 
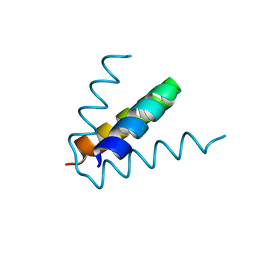 | |
6AQU
 
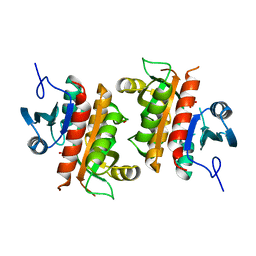 | | Crystal Structure of Plasmodium falciparum purine nucleoside phosphorylase: The M183L mutant | | Descriptor: | PHOSPHATE ION, Purine nucleoside phosphorylase | | Authors: | Harijan, R.K, Ducati, R.G, Namanja-Magliano, H.A, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2017-08-21 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Genetic resistance to purine nucleoside phosphorylase inhibition inPlasmodium falciparum.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
8PX5
 
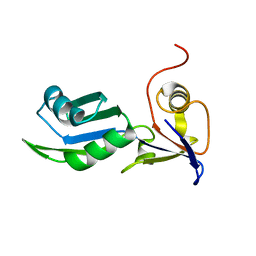 | | Structure of the RNA recognition motif (RRM) of Seb1 from S. pombe., solved at wavelength 2.75 A | | Descriptor: | Rpb7-binding protein seb1 | | Authors: | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C, Wittmann, S, Renner, M, Grimes, J.M, Wagner, A. | | Deposit date: | 2023-07-22 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
8PX9
 
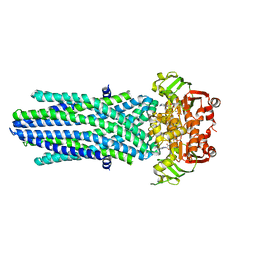 | | Structure of the antibacterial peptide ABC transporter McjD, solved at wavelength 2.75 A | | Descriptor: | MAGNESIUM ION, Microcin-J25 export ATP-binding/permease protein McjD, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER | | Authors: | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C, Bountra, K, Beis, K, Wagner, A. | | Deposit date: | 2023-07-22 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
8DJS
 
 | |
7T5T
 
 | | Structure of Thauera sp. K11 CapP | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, CapP toxin, SULFATE ION, ... | | Authors: | Lau, R.K, Corbett, K.D. | | Deposit date: | 2021-12-13 | | Release date: | 2022-09-14 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A conserved signaling pathway activates bacterial CBASS immune signaling in response to DNA damage.
Embo J., 41, 2022
|
|
6PNN
 
 | |
6AR5
 
 | |
6M7H
 
 | | Structure of calmodulin with KN93 | | Descriptor: | CALCIUM ION, Calmodulin-1, N-[2-[[[3-(4'-Chlorophenyl)-2-propenyl]methylamino]methyl]phenyl]-N-(2-hydroxyethyl)-4'-methoxybenzenesulfonamide | | Authors: | Damo, S.M, Pattanayek, R, Johnson, C.N. | | Deposit date: | 2018-08-20 | | Release date: | 2019-08-28 | | Last modified: | 2019-11-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The CaMKII inhibitor KN93-calmodulin interaction and implications for calmodulin tuning of NaV1.5 and RyR2 function.
Cell Calcium, 82, 2019
|
|
5BO4
 
 | | Structure of SOCS2:Elongin C:Elongin B from DMSO-treated crystals | | Descriptor: | Suppressor of cytokine signaling 2, Transcription elongation factor B polypeptide 1, Transcription elongation factor B polypeptide 2 | | Authors: | Gadd, M.S, Bulatov, E, Ciulli, A. | | Deposit date: | 2015-05-27 | | Release date: | 2015-07-08 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Serendipitous SAD Solution for DMSO-Soaked SOCS2-ElonginC-ElonginB Crystals Using Covalently Incorporated Dimethylarsenic: Insights into Substrate Receptor Conformational Flexibility in Cullin RING Ligases.
Plos One, 10, 2015
|
|
7WAA
 
 | | Crystal structure of MCR-1-S treated by AgNO3 | | Descriptor: | Probable phosphatidylethanolamine transferase Mcr-1, SILVER ION | | Authors: | Zhang, Q, Wang, M, Sun, H. | | Deposit date: | 2021-12-13 | | Release date: | 2022-03-16 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Re-sensitization of mcr carrying multidrug resistant bacteria to colistin by silver.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
8PX4
 
 | | Structure of the PAS domain code by the LIC_11128 gene from Leptospira interrogans serovar Copenhageni Fiocruz, solved at wavelength 3.09 A | | Descriptor: | Diguanylate cyclase | | Authors: | El Omari, K, Duman, R, Mykhaylyk, V, Orr, C, Guzzo, C.R, Owens, R.J, Wagner, A. | | Deposit date: | 2023-07-22 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Experimental phasing opportunities for macromolecular crystallography at very long wavelengths.
Commun Chem, 6, 2023
|
|
