5U0G
 
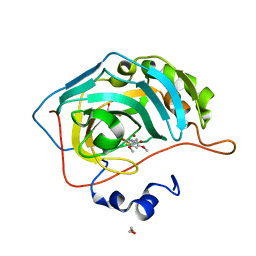 | | Identification of a New Zinc Binding Chemotype by Fragment Screening | | Descriptor: | (5R)-5-[(2,4-dimethoxyphenyl)methyl]-1,3-thiazolidine-2,4-dione, 1,2-ETHANEDIOL, Carbonic anhydrase 2, ... | | Authors: | Peat, T.S, Poulsen, S.A, Ren, B, Dolezal, O, Woods, L.A, Mujumdar, P, Chrysanthopoulos, P.K. | | Deposit date: | 2016-11-23 | | Release date: | 2017-08-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Identification of a New Zinc Binding Chemotype by Fragment Screening.
J. Med. Chem., 60, 2017
|
|
7P0X
 
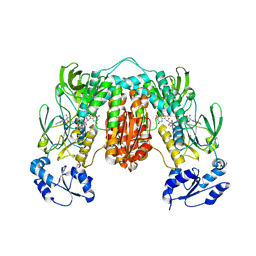 | | Crystal structure of Thioredoxin reductase from Brugia Malayi | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CHLORIDE ION, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Fata, F, Ardini, M, Silvestri, I, Gabriele, F, Cheng, Q, Arner, E.S.J, Williams, D.L. | | Deposit date: | 2021-06-30 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Biochemical and structural characterizations of thioredoxin reductase selenoproteins of the parasitic filarial nematodes Brugia malayi and Onchocerca volvulus.
Redox Biol, 51, 2022
|
|
7UUD
 
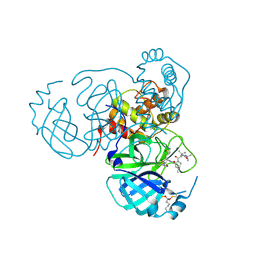 | | Structure of the SARS-CoV-2 main protease in complex with inhibitor MPI33 | | Descriptor: | (1R,2S,5S)-3-[N-(tert-butylcarbamoyl)-3-methyl-L-valyl]-N-{(2S,3R)-4-(ethylamino)-3-hydroxy-4-oxo-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Yang, K.S, Liu, W.R. | | Deposit date: | 2022-04-28 | | Release date: | 2023-01-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Novel Y-Shaped, S-O-N-O-S-Bridged Cross-Link between Three Residues C22, C44, and K61 Is Frequently Observed in the SARS-CoV-2 Main Protease.
Acs Chem.Biol., 18, 2023
|
|
7P8S
 
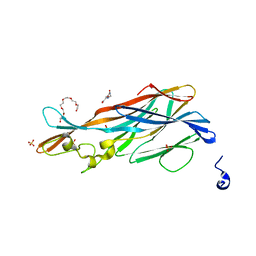 | | Crystal Structure of leukotoxin LukE from Staphylococcus aureus at 1.9 Angstrom resolution | | Descriptor: | (CARBAMOYLMETHYL-CARBOXYMETHYL-AMINO)-ACETIC ACID, HEXAETHYLENE GLYCOL, Leucotoxin LukEv, ... | | Authors: | Lambey, P, Hoh, F, Granier, S, Leyrat, C. | | Deposit date: | 2021-07-23 | | Release date: | 2022-04-06 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into recognition of chemokine receptors by Staphylococcus aureus leukotoxins.
Elife, 11, 2022
|
|
8TRP
 
 | |
8QA7
 
 | |
7P7F
 
 | | Crystal structure of phosphorylated pT220 Casein Kinase I delta (CK1d), conformation 1 | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE, ADENOSINE MONOPHOSPHATE, ... | | Authors: | Chaikuad, A, Zhubi, R, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-07-19 | | Release date: | 2022-04-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Kinase domain autophosphorylation rewires the activity and substrate specificity of CK1 enzymes.
Mol.Cell, 82, 2022
|
|
8KB1
 
 | | Crystal structure of 11JD | | Descriptor: | Beta2-microglobulin, MHC class I antigen, peptide of AIV | | Authors: | Tang, Z, Zhang, N. | | Deposit date: | 2023-08-03 | | Release date: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.46 Å) | | Cite: | The impact of micropolymorphism in Anpl-UAA on structural stability and peptide presentation.
Int.J.Biol.Macromol., 267, 2024
|
|
5C02
 
 | |
6LZB
 
 | | crystal structure of Human Methionine aminopeptidase (HsMetAP1b) in complex with AN-P2-5H-06 | | Descriptor: | 1-[(3-methoxyphenyl)methyl]-~{N}-oxidanyl-pyrrolo[2,3-b]pyridine-5-carboxamide, COBALT (II) ION, GLYCEROL, ... | | Authors: | sandeep, C.B, Addlagatta, A. | | Deposit date: | 2020-02-18 | | Release date: | 2021-02-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.29 Å) | | Cite: | Selective inhibition of Helicobacter pylori methionine aminopeptidase by azaindole hydroxamic acid derivatives: Design, synthesis, in vitro biochemical and structural studies.
Bioorg.Chem., 115, 2021
|
|
7P7G
 
 | | Crystal structure of phosphorylated pT220 Casein Kinase I delta (CK1d), conformation 2 and 3 | | Descriptor: | 1,2-ETHANEDIOL, ADENOSINE MONOPHOSPHATE, CITRIC ACID, ... | | Authors: | Chaikuad, A, Zhubi, R, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2021-07-19 | | Release date: | 2022-04-13 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Kinase domain autophosphorylation rewires the activity and substrate specificity of CK1 enzymes.
Mol.Cell, 82, 2022
|
|
5C0I
 
 | | HAL-A02 carrying RQFGPDFPTI | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, CALCIUM ION, ... | | Authors: | Rizkallah, P.J, Bulek, A.M, Cole, D.K, Sewell, A.K. | | Deposit date: | 2015-06-12 | | Release date: | 2016-05-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Hotspot autoimmune T cell receptor binding underlies pathogen and insulin peptide cross-reactivity.
J.Clin.Invest., 126, 2016
|
|
7U9N
 
 | |
7M0N
 
 | | The crystal structure of wild type PA endonuclease (A/Vietnam/1203/2004) in complex with Raltegravir | | Descriptor: | GLYCEROL, MANGANESE (II) ION, N-(4-fluorobenzyl)-5-hydroxy-1-methyl-2-(1-methyl-1-{[(5-methyl-1,3,4-oxadiazol-2-yl)carbonyl]amino}ethyl)-6-oxo-1,6-di hydropyrimidine-4-carboxamide, ... | | Authors: | Cuypers, M.G, Slavish, P.J, Yun, M.K, Dubois, R, Rankovic, Z, White, S.W. | | Deposit date: | 2021-03-11 | | Release date: | 2022-03-02 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Chemical scaffold recycling: Structure-guided conversion of an HIV integrase inhibitor into a potent influenza virus RNA-dependent RNA polymerase inhibitor designed to minimize resistance potential.
Eur.J.Med.Chem., 247, 2023
|
|
8QDV
 
 | |
8TY5
 
 | | MI-14 bound to Mpro of SARS-CoV-2 | | Descriptor: | (1R,2S,5S)-3-[(2,4-dichlorophenoxy)acetyl]-N-{(2S)-1-hydroxy-3-[(3S)-2-oxopyrrolidin-3-yl]propan-2-yl}-6,6-dimethyl-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Blankenship, L.R, Liu, W.R. | | Deposit date: | 2023-08-24 | | Release date: | 2024-08-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | MI-14 bound to SARS-CoV-2 Mpro
To Be Published
|
|
6LZC
 
 | | crystal structure of Human Methionine aminopeptidase (HsMetAP1b) in complex with KV-P2-4H-05 | | Descriptor: | COBALT (II) ION, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Sandeep, C.B, Addlagatta, A. | | Deposit date: | 2020-02-18 | | Release date: | 2021-02-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Selective inhibition of Helicobacter pylori methionine aminopeptidase by azaindole hydroxamic acid derivatives: Design, synthesis, in vitro biochemical and structural studies.
Bioorg.Chem., 115, 2021
|
|
7XRZ
 
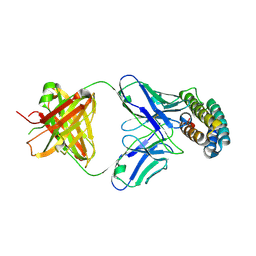 | | Crystal structure of BRIL and SRP2070_Fab complex | | Descriptor: | IGG HEAVY CHAIN, IGG LIGHT CHAIN, Soluble cytochrome b562 | | Authors: | Suzuki, M, Miyagi, H, Yasunaga, M, Asada, H, Iwata, S, Saito, J. | | Deposit date: | 2022-05-12 | | Release date: | 2023-05-10 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural insight into an anti-BRIL Fab as a G-protein-coupled receptor crystallization chaperone.
Acta Crystallogr D Struct Biol, 79, 2023
|
|
8QAN
 
 | |
5G4Z
 
 | | Structural basis for carboxylic acid recognition by a Cache chemosensory domain. | | Descriptor: | Methyl-accepting chemotaxis sensory transducer with Cache sensor, TRIETHYLENE GLYCOL, UNKNOWN LIGAND | | Authors: | Brewster, J, McKellar, J.L.O, Newman, J, Peat, T.S, Gerth, M.L. | | Deposit date: | 2016-05-18 | | Release date: | 2017-03-29 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural basis for ligand recognition by a Cache chemosensory domain that mediates carboxylate sensing in Pseudomonas syringae.
Sci Rep, 6, 2016
|
|
7P7H
 
 | |
7MQ2
 
 | |
8QAP
 
 | | Crystal Structure of the first bromodomain of BRD4 in complex with acetyl-pyrrole derivative compound 2 | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-(3,6,6-trimethyl-4-oxidanylidene-5,7-dihydro-2~{H}-isoindol-1-yl)-1,3-thiazol-2-yl]guanidine, Bromodomain-containing protein 4 | | Authors: | Dalle Vedove, A, Cazzanelli, G, Lolli, G. | | Deposit date: | 2023-08-23 | | Release date: | 2024-09-11 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Dual BET/BRPF1 inhibitors induce morphological alteration and cellular death in various human cancer cell lines
To be published
|
|
5G5V
 
 | | Crystal structure of T. brucei PDE-B1 catalytic domain with inhibitor NPD-038 | | Descriptor: | (4AS,8AR)-4-(3-{4-[(3R)-3-HYDROXYPYRROLIDINE-1-, 1,2-ETHANEDIOL, FORMIC ACID, ... | | Authors: | Singh, A.K, Brown, D.G. | | Deposit date: | 2016-06-06 | | Release date: | 2018-03-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Targeting a Subpocket in Trypanosoma brucei Phosphodiesterase B1 (TbrPDEB1) Enables the Structure-Based Discovery of Selective Inhibitors with Trypanocidal Activity.
J. Med. Chem., 61, 2018
|
|
7Y86
 
 | | CcpS mutant | | Descriptor: | UPF0297 protein A7J08_00425 | | Authors: | Tang, J.S, Ran, T.T, Wang, W.W, Fan, H.J. | | Deposit date: | 2022-06-22 | | Release date: | 2023-05-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A link between STK signalling and capsular polysaccharide synthesis in Streptococcus suis.
Nat Commun, 14, 2023
|
|
