3WFA
 
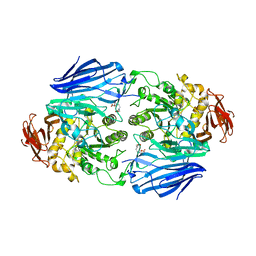 | | Catalytic role of the calcium ion in GH97 inverting glycoside hydrolase | | Descriptor: | Alpha-glucosidase, SODIUM ION, {[-(BIS-CARBOXYMETHYL-AMINO)-ETHYL]-CARBOXYMETHYL-AMINO}-ACETIC ACID | | Authors: | Okuyama, M, Yoshida, T, Hondoh, H, Mori, H, Yao, M, Kimura, A. | | Deposit date: | 2013-07-18 | | Release date: | 2014-07-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Catalytic role of the calcium ion in GH97 inverting glycoside hydrolase
To be Published
|
|
6PLA
 
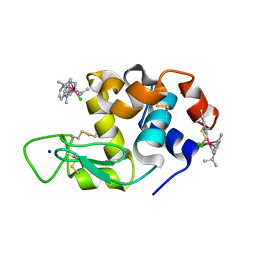 | | Adducts formed after 1 month in the reaction of dichlorido(1,3-dimethylbenzimidazol-2-ylidene)(eta6-p-cymene)osmium(II) with HEWL | | Descriptor: | Lysozyme, OSMIUM ION, SODIUM ION, ... | | Authors: | Sullivan, M.P, Hartinger, C.G, Goldstone, D.C. | | Deposit date: | 2019-06-30 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Probing the Paradigm of Promiscuity for N-Heterocyclic Carbene Complexes and their Protein Adduct Formation.
Angew.Chem.Int.Ed.Engl., 2021
|
|
6TPD
 
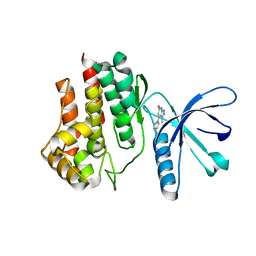 | | Fragment-based discovery of pyrazolopyridones as JAK1 inhibitors with excellent subtype selectivity | | Descriptor: | 3-methyl-4-phenyl-2,7-dihydropyrazolo[3,4-b]pyridin-6-one, Tyrosine-protein kinase JAK2 | | Authors: | Hansen, B.B, Jepsen, T.J, Larsen, M, Sindet, R, Vifian, T, Burhardt, M.N, Larsen, J, Seitzberg, J.G, Carnerup, M.A, Jerre, A, Moelck, C, Rai, S, Nasipireddy, V.R, Ritzen, A. | | Deposit date: | 2019-12-13 | | Release date: | 2020-06-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Fragment-Based Discovery of Pyrazolopyridones as JAK1 Inhibitors with Excellent Subtype Selectivity.
J.Med.Chem., 63, 2020
|
|
6PMP
 
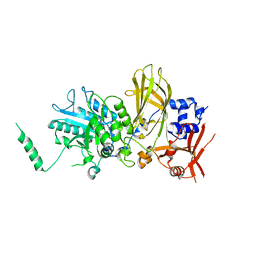 | |
6VHS
 
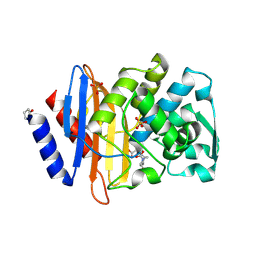 | | Crystal structure of CTX-M-14 in complex with beta-lactamase inhibitor ETX1317 | | Descriptor: | (2R)-({[(3R,6S)-6-carbamoyl-1-formyl-4-methyl-1,2,3,6-tetrahydropyridin-3-yl]amino}oxy)(fluoro)acetic acid, Beta-lactamase, PHOSPHATE ION | | Authors: | Sacco, M.D, Chen, Y. | | Deposit date: | 2020-01-10 | | Release date: | 2020-08-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.28 Å) | | Cite: | Discovery of an Orally Available Diazabicyclooctane Inhibitor (ETX0282) of Class A, C, and D Serine beta-Lactamases.
J.Med.Chem., 63, 2020
|
|
3JUG
 
 | |
6TJ0
 
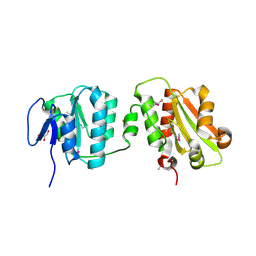 | | Crystal structure of the bacterial cellulose secretion regulator BcsE, residues 217-523, with bound c-di-GMP. | | Descriptor: | 9,9'-[(2R,3R,3aS,5S,7aR,9R,10R,10aS,12S,14aR)-3,5,10,12-tetrahydroxy-5,12-dioxidooctahydro-2H,7H-difuro[3,2-d:3',2'-j][1,3,7,9,2,8]tetraoxadiphosphacyclododecine-2,9-diyl]bis(2-amino-1,9-dihydro-6H-purin-6-one), Bacterial cellulose synthesis subunit E, GLYCEROL | | Authors: | Zouhir, S, Abidi, W, Krasteva, P.V. | | Deposit date: | 2019-11-23 | | Release date: | 2020-07-29 | | Last modified: | 2020-08-26 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and Multitasking of the c-di-GMP-Sensing Cellulose Secretion Regulator BcsE.
Mbio, 11, 2020
|
|
3WW4
 
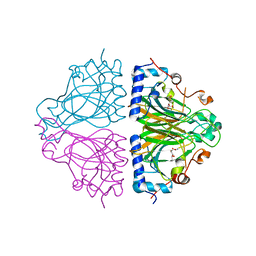 | | X-ray structures of Cellulomonas parahominis L-ribose isomerase with L-allose | | Descriptor: | L-allose, L-ribose isomerase, MANGANESE (II) ION, ... | | Authors: | Terami, Y, Yoshida, H, Takata, G, Kamitori, S. | | Deposit date: | 2014-06-13 | | Release date: | 2015-04-29 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Essentiality of tetramer formation of Cellulomonas parahominis L-ribose isomerase involved in novel L-ribose metabolic pathway.
Appl.Microbiol.Biotechnol., 99, 2015
|
|
3K91
 
 | | Polysulfane Bridge in Cu-Zn Superoxide Dismutase | | Descriptor: | PENTASULFIDE-SULFUR, Superoxide dismutase [Cu-Zn] | | Authors: | You, Z, Cao, X, Taylor, A.B, Hart, P.J, Levine, R.L. | | Deposit date: | 2009-10-15 | | Release date: | 2010-01-19 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Characterization of a covalent polysulfane bridge in copper-zinc superoxide dismutase .
Biochemistry, 49, 2010
|
|
6PEY
 
 | | MTHFR with mutation Asp120Ala | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, Methylenetetrahydrofolate reductase | | Authors: | Gallagher, E.L, Gurney, L.A, Frasco, F.G, Trimmer, E, Bolen, R.L, Collins, K, Garland, E, Halloran, J, Handley-Pendelton, J, Hernandez, V, Leffler, S, Perez, A, Soares, A, Stojanoff, V, Williams, D. | | Deposit date: | 2019-06-21 | | Release date: | 2019-09-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Examination of Asp120Ala a Chemically Important Novel Mutation in the Enzyme Mthylenetetrahydrofolate Reductase
To be published
|
|
6TMN
 
 | | Structures of two thermolysin-inhibitor complexes that differ by a single hydrogen bond | | Descriptor: | CALCIUM ION, N-[(2R,4S)-4-hydroxy-2-(2-methylpropyl)-4-oxido-7-oxo-9-phenyl-3,8-dioxa-6-aza-4-phosphanonan-1-oyl]-L-leucine, THERMOLYSIN, ... | | Authors: | Tronrud, D.E, Holden, H.M, Matthews, B.W. | | Deposit date: | 1987-06-29 | | Release date: | 1989-01-09 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of two thermolysin-inhibitor complexes that differ by a single hydrogen bond.
Science, 235, 1987
|
|
6T4T
 
 | | ROR(gamma)t ligand binding domain in complex with 20-alpha-hydroxycholesterol and allosteric ligand FM26 | | Descriptor: | (3alpha,8alpha)-cholest-5-ene-3,20-diol, 4-[(~{E})-[3-[2-chloranyl-6-(trifluoromethyl)phenyl]-5-(1~{H}-pyrrol-3-yl)-1,2-oxazol-4-yl]methylideneamino]benzoic acid, GLYCEROL, ... | | Authors: | de Vries, R.M.J.M, Meijer, F.A, Brunsveld, L. | | Deposit date: | 2019-10-15 | | Release date: | 2020-11-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | Cooperativity between the orthosteric and allosteric ligand binding sites of ROR gamma t.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
6PLB
 
 | | Adducts formed after 1 month in the reaction of dichlorido(1,3-dimethylbenzimida zol-2-ylidene)(eta5-pentamethylcyclopentadienyl)iridium(III) with HEWL | | Descriptor: | 2-(1-chloranyl-2,3,4,5,6-pentamethyl-1$l^{7}-iridapentacyclo[2.2.0.0^{1,3}.0^{1,5}.0^{2,6}]hexan-1-yl)-1,3-dimethyl-benzimidazole, Lysozyme, SODIUM ION | | Authors: | Sullivan, M.P, Hartinger, C.G, Goldstone, D.C. | | Deposit date: | 2019-06-30 | | Release date: | 2020-11-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Probing the Paradigm of Promiscuity for N-Heterocyclic Carbene Complexes and their Protein Adduct Formation.
Angew.Chem.Int.Ed.Engl., 2021
|
|
6VO5
 
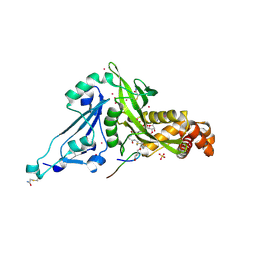 | | Crystal structure of Human histone acetytransferas 1 (HAT1) in complex with isobutryl-COA and K12A mutant variant of histone H4 | | Descriptor: | ACETATE ION, GLYCEROL, Histone H4, ... | | Authors: | Halabelian, L, Zeng, H, Dong, A, Loppnau, P, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-01-30 | | Release date: | 2020-03-11 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of Human histone acetytransferas 1 (HAT1) in complex with isobutryl-COA and K12A mutant variant of histone H4
to be published
|
|
6TOR
 
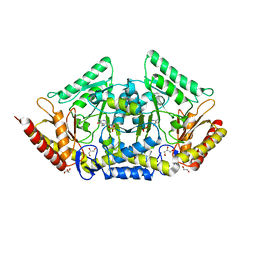 | | human O-phosphoethanolamine phospho-lyase | | Descriptor: | 4'-DEOXY-4'-AMINOPYRIDOXAL-5'-PHOSPHATE, Ethanolamine-phosphate phospho-lyase, GLYCEROL | | Authors: | Vettraino, C, Donini, S, Parisini, E. | | Deposit date: | 2019-12-11 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural characterization of human O-phosphoethanolamine phospho-lyase.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
4H2L
 
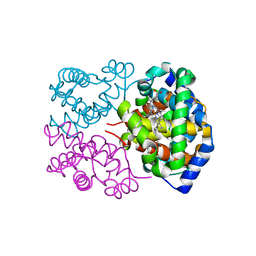 | | Deer mouse hemoglobin in hydrated format | | Descriptor: | Alpha-globin, Beta globin, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Inoguchi, N, Oshlo, J.R, Natarajan, C, Weber, R.E, Fago, A, Storz, J.F, Moriyama, H. | | Deposit date: | 2012-09-12 | | Release date: | 2013-04-10 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.779 Å) | | Cite: | Deer mouse hemoglobin exhibits a lowered oxygen affinity owing to mobility of the E helix.
Acta Crystallogr.,Sect.F, 69, 2013
|
|
6VRS
 
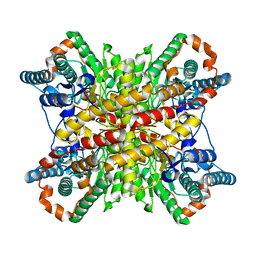 | | Single particle reconstruction of glucose isomerase from Streptomyces rubiginosus based on data acquired in the presence of substantial aberrations | | Descriptor: | MANGANESE (II) ION, xylose isomerase | | Authors: | Bromberg, R, Guo, Y, Borek, D, Otwinowski, Z. | | Deposit date: | 2020-02-09 | | Release date: | 2020-02-19 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | High-resolution cryo-EM reconstructions in the presence of substantial aberrations
Iucrj, 7, 2020
|
|
4GR2
 
 | | Structure of AtRbcX1 from Arabidopsis thaliana. | | Descriptor: | AtRbcX1 | | Authors: | Golik, P, Grudnik, P, Kolesinski, P, Dubin, G, Szczepaniak, A. | | Deposit date: | 2012-08-24 | | Release date: | 2013-01-30 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Insights into eukaryotic Rubisco assembly - Crystal structures of RbcX chaperones from Arabidopsis thaliana.
Biochim.Biophys.Acta, 1830, 2013
|
|
6T5U
 
 | | KRasG12C ligand complex | | Descriptor: | 1-[(7R)-16-chloro-15-(5-methyl-1H-indazol-4-yl)-9-oxa-2,5,12-triazatetracyclo[8.8.0.02,7.013,18]octadeca-1(10),11,13,15,17-pentaen-5-yl]prop-2-en-1-one, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Phillips, C. | | Deposit date: | 2019-10-17 | | Release date: | 2020-02-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.72 Å) | | Cite: | Structure-Based Design and Pharmacokinetic Optimization of Covalent Allosteric Inhibitors of the Mutant GTPase KRASG12C.
J.Med.Chem., 63, 2020
|
|
3JXJ
 
 | |
3D17
 
 | | A triply ligated crystal structure of relaxed state human hemoglobin | | Descriptor: | CARBON MONOXIDE, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Safo, M.K, Musayev, F.N, Jenkins, J, Abraham, D.J. | | Deposit date: | 2008-05-05 | | Release date: | 2008-06-03 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A triply ligated crystal structure of relaxed state human hemoglobin
TO BE PUBLISHED
|
|
6P9A
 
 | | HIV-1 Protease multiple mutant PRS5B with Darunavir | | Descriptor: | (3R,3AS,6AR)-HEXAHYDROFURO[2,3-B]FURAN-3-YL(1S,2R)-3-[[(4-AMINOPHENYL)SULFONYL](ISOBUTYL)AMINO]-1-BENZYL-2-HYDROXYPROPYLCARBAMATE, HIV-1 Protease, PHOSPHATE ION | | Authors: | Kneller, D.W, Agniswamy, J, Weber, I.T. | | Deposit date: | 2019-06-10 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Highly drug-resistant HIV-1 protease reveals decreased intra-subunit interactions due to clusters of mutations.
Febs J., 287, 2020
|
|
6TC4
 
 | | AA13 Lytic polysaccharide monooxygenase from Aspergillus oryzae measured with SSX | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, AoAA13, CHLORIDE ION, ... | | Authors: | Tandrup, T, Muderspach, S.J, Frandsen, K.E.H, Santoni, G, Poulsen, J.C.N, Lo Leggio, L. | | Deposit date: | 2019-11-05 | | Release date: | 2020-03-18 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Further structural studies of the lytic polysaccharide monooxygenase AoAA13 belonging to the starch-active AA13 family
Amylase, 3(1), 2019
|
|
6PBL
 
 | |
6T96
 
 | | Photorhabdus laumondii subsp. laumondii lectin PLL3 | | Descriptor: | Lectin PLL3, SODIUM ION | | Authors: | Melicher, F, Houser, J, Fujdiarova, E, Faltinek, L, Wimmerova, M. | | Deposit date: | 2019-10-26 | | Release date: | 2019-12-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Lectin PLL3, a Novel Monomeric Member of the Seven-Bladed beta-Propeller Lectin Family.
Molecules, 24, 2019
|
|
