7PKB
 
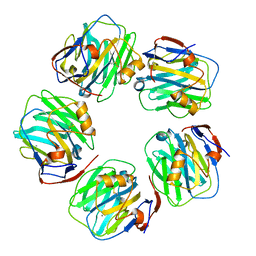 | | C-reactive protein pentamer at pH 7.5 | | Descriptor: | C-reactive protein, CALCIUM ION | | Authors: | Noone, D.P, Sharp, T.H. | | Deposit date: | 2021-08-25 | | Release date: | 2021-12-22 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-Electron Microscopy and Biochemical Analysis Offer Insights Into the Effects of Acidic pH, Such as Occur During Acidosis, on the Complement Binding Properties of C-Reactive Protein.
Front Immunol, 12, 2021
|
|
7PKF
 
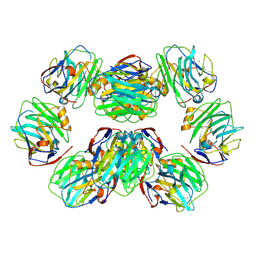 | | C-reactive protein decamer at pH 5 | | Descriptor: | C-reactive protein, CALCIUM ION | | Authors: | Noone, D.P, Sharp, T.H. | | Deposit date: | 2021-08-25 | | Release date: | 2021-12-22 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-Electron Microscopy and Biochemical Analysis Offer Insights Into the Effects of Acidic pH, Such as Occur During Acidosis, on the Complement Binding Properties of C-Reactive Protein.
Front Immunol, 12, 2021
|
|
7PK9
 
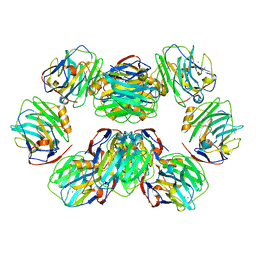 | | C-reactive protein decamer at pH 7.5 | | Descriptor: | C-reactive protein, CALCIUM ION | | Authors: | Noone, D.P, Sharp, T.H. | | Deposit date: | 2021-08-25 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Cryo-Electron Microscopy and Biochemical Analysis Offer Insights Into the Effects of Acidic pH, Such as Occur During Acidosis, on the Complement Binding Properties of C-Reactive Protein.
Front Immunol, 12, 2021
|
|
7PKD
 
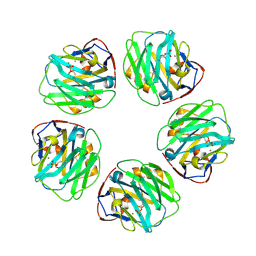 | | C-reactive protein decamer at pH 7.5 with phosphocholine ligand | | Descriptor: | C-reactive protein, CALCIUM ION, PHOSPHOCHOLINE | | Authors: | Noone, D.P, Sharp, T.H. | | Deposit date: | 2021-08-25 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-Electron Microscopy and Biochemical Analysis Offer Insights Into the Effects of Acidic pH, Such as Occur During Acidosis, on the Complement Binding Properties of C-Reactive Protein.
Front Immunol, 12, 2021
|
|
7RIB
 
 | | Griffithsin mutant Y28F/Y68F/Y110F | | Descriptor: | Griffithsin, SULFATE ION, alpha-D-mannopyranose | | Authors: | Zhao, G, Sun, J, Bewley, C.A. | | Deposit date: | 2021-07-19 | | Release date: | 2022-05-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | C 3 -Symmetric Aromatic Core of Griffithsin Is Essential for Potent Anti-HIV Activity.
Acs Chem.Biol., 17, 2022
|
|
7PKE
 
 | | C-reactive protein pentamer at pH 7.5 with phosphocholine ligand | | Descriptor: | C-reactive protein, CALCIUM ION, PHOSPHOCHOLINE | | Authors: | Noone, D.P, Sharp, T.H. | | Deposit date: | 2021-08-25 | | Release date: | 2021-12-22 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-Electron Microscopy and Biochemical Analysis Offer Insights Into the Effects of Acidic pH, Such as Occur During Acidosis, on the Complement Binding Properties of C-Reactive Protein.
Front Immunol, 12, 2021
|
|
7PKH
 
 | | C-reactive protein decamer at pH 5 with phosphocholine ligand | | Descriptor: | C-reactive protein, CALCIUM ION, PHOSPHOCHOLINE | | Authors: | Noone, D.P, Sharp, T.H. | | Deposit date: | 2021-08-25 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Cryo-Electron Microscopy and Biochemical Analysis Offer Insights Into the Effects of Acidic pH, Such as Occur During Acidosis, on the Complement Binding Properties of C-Reactive Protein.
Front Immunol, 12, 2021
|
|
7PKG
 
 | | C-reactive protein pentamer at pH 5 | | Descriptor: | C-reactive protein, CALCIUM ION | | Authors: | Noone, D.P, Sharp, T.H. | | Deposit date: | 2021-08-25 | | Release date: | 2021-12-22 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Cryo-Electron Microscopy and Biochemical Analysis Offer Insights Into the Effects of Acidic pH, Such as Occur During Acidosis, on the Complement Binding Properties of C-Reactive Protein.
Front Immunol, 12, 2021
|
|
8FX4
 
 | | GC-C-Hsp90-Cdc37 regulatory complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Guanylyl cyclase C, Heat shock protein HSP 90-beta, ... | | Authors: | Caveney, N.A, Garcia, K.C. | | Deposit date: | 2023-01-23 | | Release date: | 2023-07-12 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural insight into guanylyl cyclase receptor hijacking of the kinase-Hsp90 regulatory mechanism.
Elife, 12, 2023
|
|
8VFO
 
 | | Crystal Structure of L117G Variant of D-Dopachrome Tautomerase (D-DT) | | Descriptor: | CITRIC ACID, D-dopachrome decarboxylase | | Authors: | Parkins, A, Pilien, A, Thompson, M.C, Pantouris, G. | | Deposit date: | 2023-12-21 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | The C-terminal Region of D-DT Regulates Molecular Recognition for Protein-Ligand Complexes.
J.Med.Chem., 67, 2024
|
|
8VG8
 
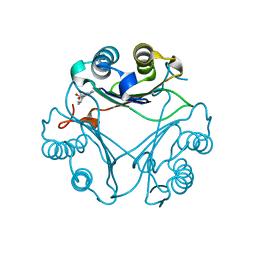 | | Crystal Structure of T115A Variant of D-Dopachrome Tautomerase (D-DT) Bound to 4CPPC | | Descriptor: | 4-(3-carboxyphenyl)pyridine-2,5-dicarboxylic acid, D-dopachrome decarboxylase | | Authors: | Parkins, A, Pilien, A, Wolff, A, Thompson, M.C, Pantouris, G. | | Deposit date: | 2023-12-23 | | Release date: | 2024-05-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.33 Å) | | Cite: | The C-terminal Region of D-DT Regulates Molecular Recognition for Protein-Ligand Complexes.
J.Med.Chem., 67, 2024
|
|
6UL7
 
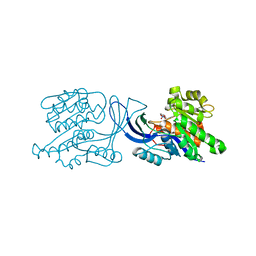 | | Structure of human ketohexokinase-C in complex with fructose, NO3, and osthole | | Descriptor: | 7-methoxy-8-(3-methylbut-2-enyl)chromen-2-one, Ketohexokinase, NITRATE ION, ... | | Authors: | Gasper, W.C, Gardner, S, Allen, K.N, Tolan, D.R. | | Deposit date: | 2019-10-07 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structure of human ketohexokinase-C in complex with fructose, NO3, and osthole
To be Published
|
|
8Q1W
 
 | |
6XA3
 
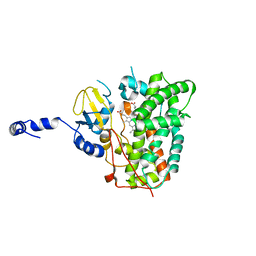 | | Structure of the ligand free P450 monooxygenase TamI | | Descriptor: | PROTOPORPHYRIN IX CONTAINING FE, TamI | | Authors: | Newmister, S.A, Srivastava, K.R, Espinoza, R.V, Haatveit, K.C, Khatri, Y, Martini, R.M, Garcia-Borras, M, Podust, L.M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2020-06-03 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.96 Å) | | Cite: | Molecular Basis of Iterative C─H Oxidation by TamI, a Multifunctional P450 monooxygenase from the Tirandamycin Biosynthetic Pathway.
Acs Catalysis, 10, 2020
|
|
7PVX
 
 | |
8R5D
 
 | | Crystal structure of human TRIM7 PRYSPRY domain | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase TRIM7, MALONIC ACID | | Authors: | Munoz Sosa, C.J, Kraemer, A, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-11-16 | | Release date: | 2024-01-17 | | Last modified: | 2024-07-31 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | A C-Degron Structure-Based Approach for the Development of Ligands Targeting the E3 Ligase TRIM7.
Acs Chem.Biol., 19, 2024
|
|
7QAN
 
 | | Cytochrome P450 Enzyme AbyV | | Descriptor: | 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CHLORIDE ION, Cytochrome P450, ... | | Authors: | Parnell, A.E, Back, C.R, Race, P.R. | | Deposit date: | 2021-11-17 | | Release date: | 2022-11-30 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.01 Å) | | Cite: | The Role of Cytochrome P450 AbyV in the Final Stages of Abyssomicin C Biosynthesis.
Angew.Chem.Int.Ed.Engl., 62, 2023
|
|
3E73
 
 | | Crystal Structure of Human LanCL1 complexed with GSH | | Descriptor: | GLUTATHIONE, LanC-like protein 1, ZINC ION | | Authors: | Zhang, W, Zhu, G, Li, X, Rao, Z, Zhang, C. | | Deposit date: | 2008-08-17 | | Release date: | 2009-06-30 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of human lanthionine synthetase C-like protein 1 and its interaction with Eps8 and glutathione
Genes Dev., 23, 2009
|
|
8R3L
 
 | | Influenza A/H7N9 polymerase in pre-initiation state, intermediate conformation (I) with PB2-C(I), ENDO(T), and Pol II pS5 CTD peptide mimic bound in site 1A/2A | | Descriptor: | 3' vRNA end (51-mer vRNA loop), Polymerase acidic protein, Polymerase basic protein 2, ... | | Authors: | Arragain, B, Cusack, S. | | Deposit date: | 2023-11-09 | | Release date: | 2024-02-21 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | The host RNA polymerase II C-terminal domain is the anchor for replication of the influenza virus genome.
Nat Commun, 15, 2024
|
|
8T3I
 
 | | Crystal structure of mutant exfoliative toxin C (ExhC) from Mammaliicoccus sciuri | | Descriptor: | Exfoliative toxin C | | Authors: | Gismene, C, Calil, F.A, Hernandez Gonzalez, J.E, Ziem Nascimento, A.F, Santisteban, A.R.N, Arni, R.K, Barros Mariutti, R. | | Deposit date: | 2023-06-07 | | Release date: | 2023-11-08 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Necrotic activity of ExhC from Mammaliicoccus sciuri is mediated by specific amino acid residues.
Int.J.Biol.Macromol., 254, 2024
|
|
3F2Z
 
 | | Crystal structure of the C-terminal domain of a chitobiase (BF3579) from Bacteroides fragilis, Northeast Structural Genomics Consortium Target BfR260B | | Descriptor: | uncharacterized protein BF3579 | | Authors: | Forouhar, F, Lew, S, Seetharaman, J, Janjua, H, Xiao, R, Foote, E.L, Ciccosanti, C, Lee, D, Nair, R, Everett, J.K, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2008-10-30 | | Release date: | 2008-11-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Crystal structure of the C-terminal domain of a chitobiase (BF3579) from Bacteroides fragilis, Northeast Structural Genomics Consortium Target BfR260B
To be Published
|
|
8R4Z
 
 | |
4Y5H
 
 | |
5MPT
 
 | | Structure of the citrinin polyketide synthase CMeT domain | | Descriptor: | 1,2-ETHANEDIOL, Citrinin polyketide synthase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Herbst, D.A, Storm, P.A, Townsend, C.A, Maier, T. | | Deposit date: | 2016-12-19 | | Release date: | 2017-02-22 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.648 Å) | | Cite: | Functional and Structural Analysis of Programmed C-Methylation in the Biosynthesis of the Fungal Polyketide Citrinin.
Cell Chem Biol, 24, 2017
|
|
6MZJ
 
 | | Germline VRC01 antibody recognition of a modified clade C HIV-1 envelope trimer, 2 Fabs bound, sharpened map | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 426c DS-SOSIP D3, ... | | Authors: | Borst, A.J, Weidle, C.E, Gray, M.D, Frenz, B, Snijder, J, Joyce, M.G, Georgiev, I.S, Stewart-Jones, G.B.E, Kwong, P.D, McGuire, A.T, DiMaio, F, Stamatatos, L, Pancera, M, Veesler, D. | | Deposit date: | 2018-11-05 | | Release date: | 2018-11-14 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | Germline VRC01 antibody recognition of a modified clade C HIV-1 envelope trimer and a glycosylated HIV-1 gp120 core.
Elife, 7, 2018
|
|
