4F43
 
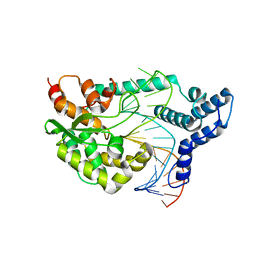 | |
4F41
 
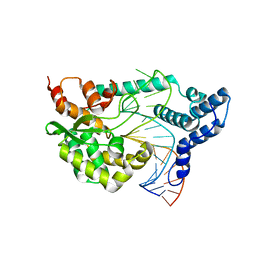 | |
3K4G
 
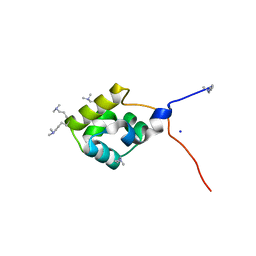 | |
1Z3E
 
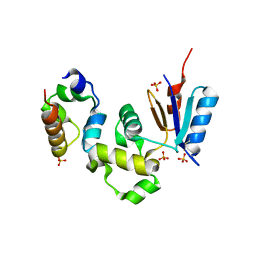 | | Crystal Structure of Spx in Complex with the C-terminal Domain of the RNA Polymerase Alpha Subunit | | Descriptor: | DNA-directed RNA polymerase alpha chain, Regulatory protein spx, SULFATE ION | | Authors: | Newberry, K.J, Nakano, S, Zuber, P, Brennan, R.G. | | Deposit date: | 2005-03-11 | | Release date: | 2005-10-11 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the Bacillus subtilis anti-alpha, global transcriptional regulator, Spx, in complex with the {alpha} C-terminal domain of RNA polymerase
Proc.Natl.Acad.Sci.Usa, 102, 2005
|
|
119D
 
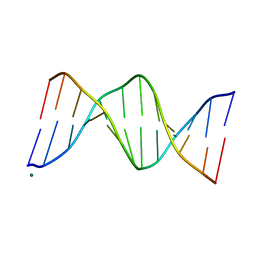 | |
1EVP
 
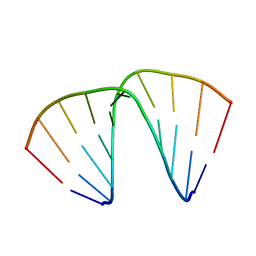 | |
5CO0
 
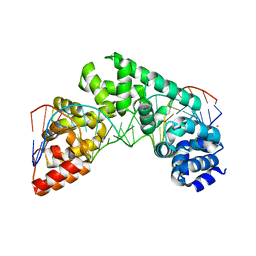 | | Crystal Structure of the MTERF1 Y288A substitution bound to the termination sequence. | | Descriptor: | DNA (5'-D(*AP*TP*TP*AP*CP*CP*GP*GP*GP*CP*TP*CP*TP*GP*CP*CP*AP*TP*CP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*GP*AP*TP*GP*GP*CP*AP*GP*AP*GP*CP*CP*CP*GP*GP*TP*AP*AP*T)-3'), POTASSIUM ION, ... | | Authors: | Byrnes, J, Hauser, K, Norona, L, Mejia, E, Simmerling, C, Garcia-Diaz, M. | | Deposit date: | 2015-07-18 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Base Flipping by MTERF1 Can Accommodate Multiple Conformations and Occurs in a Stepwise Fashion.
J.Mol.Biol., 428, 2016
|
|
6T22
 
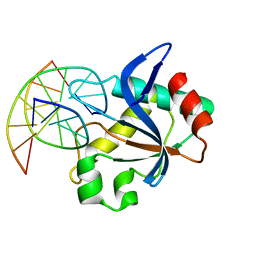 | | N-terminal domain of EcoKMcrA restriction endonuclease (NEco) in complex with T5hmCGA target sequence | | Descriptor: | DNA (5'-D(*GP*AP*AP*TP*(5HC)P*GP*AP*TP*GP*A)-3'), DNA (5'-D(*TP*CP*AP*TP*(5HC)P*GP*AP*TP*TP*C)-3'), EcoKMcrA modification dependent restriction endonuclease | | Authors: | Slyvka, A, Zagorskaite, E, Czapinska, H, Sasnauskas, G, Bochtler, M. | | Deposit date: | 2019-10-07 | | Release date: | 2019-10-30 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | Crystal structure of the EcoKMcrA N-terminal domain (NEco): recognition of modified cytosine bases without flipping.
Nucleic Acids Res., 47, 2019
|
|
5CRJ
 
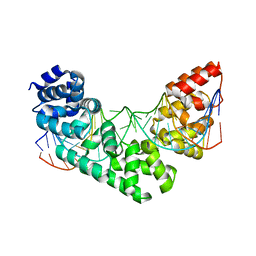 | | Crystal Structure of the MTERF1 F322A substitution bound to the termination sequence. | | Descriptor: | DNA (5'-D(*AP*TP*TP*AP*CP*CP*GP*GP*GP*CP*TP*CP*TP*GP*CP*CP*AP*TP*CP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*GP*AP*TP*GP*GP*CP*AP*GP*AP*GP*CP*CP*CP*GP*GP*TP*AP*AP*T)-3'), Transcription termination factor 1, ... | | Authors: | Byrnes, J, Hauser, K, Norona, L, Mejia, E, Simmerling, C, Garcia-Diaz, M. | | Deposit date: | 2015-07-23 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.59 Å) | | Cite: | Base Flipping by MTERF1 Can Accommodate Multiple Conformations and Occurs in a Stepwise Fashion.
J.Mol.Biol., 428, 2016
|
|
6PWX
 
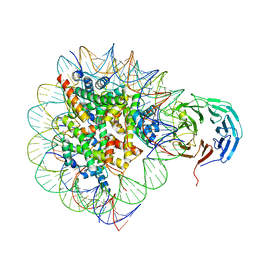 | | Cryo-EM structure of RbBP5 bound to the nucleosome | | Descriptor: | DNA (146-MER), Histone H2A type 1, Histone H2B 1.1, ... | | Authors: | Park, S.H, Ayoub, A, Lee, Y.T, Xu, J, Zhang, W, Zhang, B, Zhang, Y, Cianfrocco, M.A, Su, M, Dou, Y, Cho, U. | | Deposit date: | 2019-07-23 | | Release date: | 2019-12-18 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Cryo-EM structure of the human MLL1 core complex bound to the nucleosome.
Nat Commun, 10, 2019
|
|
6T93
 
 | | Nucleosome with OCT4-SOX2 motif at SHL-6 | | Descriptor: | DNA (153-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Michael, A.K, Kempf, G, Cavadini, S, Bunker, R.D, Thoma, N.H. | | Deposit date: | 2019-10-25 | | Release date: | 2020-05-06 | | Last modified: | 2024-05-22 | | Method: | ELECTRON MICROSCOPY (3.49 Å) | | Cite: | Mechanisms of OCT4-SOX2 motif readout on nucleosomes.
Science, 368, 2020
|
|
5CRK
 
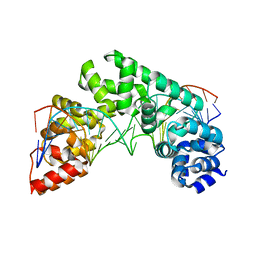 | | Crystal Structure of the MTERF1 F243A substitution bound to the termination sequence. | | Descriptor: | DNA (5'-D(*AP*TP*TP*AP*CP*CP*GP*GP*GP*CP*TP*CP*TP*GP*CP*CP*AP*TP*CP*TP*TP*A)-3'), DNA (5'-D(*TP*AP*AP*GP*AP*TP*GP*GP*CP*AP*GP*AP*GP*CP*CP*CP*GP*GP*TP*AP*AP*T)-3'), Transcription termination factor 1, ... | | Authors: | Byrnes, J, Hauser, K, Norona, L, Mejia, E, Simmerling, C, Garcia-Diaz, M. | | Deposit date: | 2015-07-23 | | Release date: | 2015-11-25 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Base Flipping by MTERF1 Can Accommodate Multiple Conformations and Occurs in a Stepwise Fashion.
J.Mol.Biol., 428, 2016
|
|
8VFZ
 
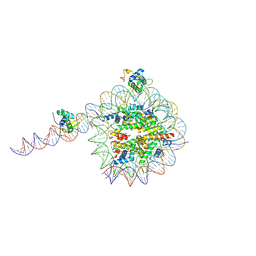 | |
8VG1
 
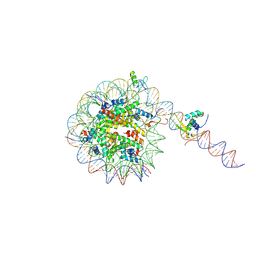 | |
8VFX
 
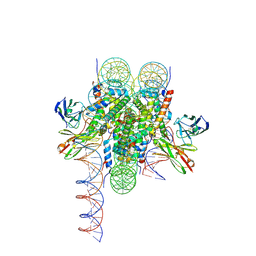 | | Cryo-EM structure of 186bp ALBN1 nucleosome aided by scFv | | Descriptor: | DNA (158-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Zhou, B.R, Bai, Y. | | Deposit date: | 2023-12-22 | | Release date: | 2024-08-07 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (2.65 Å) | | Cite: | Structural insights into the cooperative nucleosome recognition and chromatin opening by FOXA1 and GATA4.
Mol.Cell, 84, 2024
|
|
8VG0
 
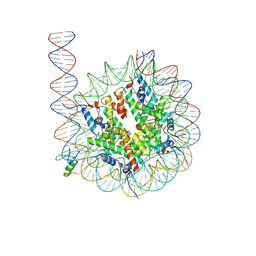 | | Cryo-EM structure of GATA4 in complex with ALBN1 nucleosome | | Descriptor: | DNA (159-MER), Histone H2A type 1-B/E, Histone H2B type 1-J, ... | | Authors: | Zhou, B.R, Bai, Y. | | Deposit date: | 2023-12-22 | | Release date: | 2024-08-07 | | Last modified: | 2024-09-11 | | Method: | ELECTRON MICROSCOPY (3.07 Å) | | Cite: | Structural insights into the cooperative nucleosome recognition and chromatin opening by FOXA1 and GATA4.
Mol.Cell, 84, 2024
|
|
7K5Y
 
 | |
9BNA
 
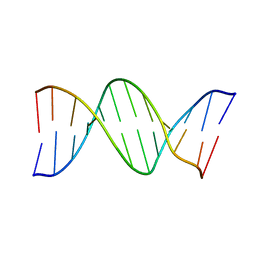 | |
2JNW
 
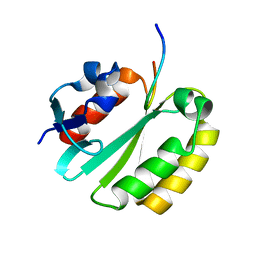 | | Solution structure of a ERCC1-XPA heterodimer | | Descriptor: | DNA excision repair protein ERCC-1, DNA-repair protein complementing XP-A cells | | Authors: | Tsodikov, O.V, Ivanov, D, Orelli, B, Staresincic, L, Scharer, O.D, Wagner, G. | | Deposit date: | 2007-02-07 | | Release date: | 2007-10-30 | | Last modified: | 2023-12-20 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the recruitment of ERCC1-XPF to nucleotide excision repair complexes by XPA
Embo J., 26, 2007
|
|
5KW6
 
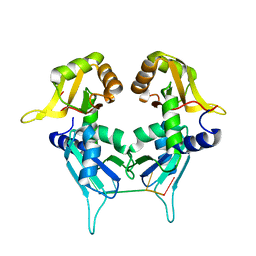 | | Two Tandem RRM Domains of PUF60 Bound to an AdML Pre-mRNA 3' Splice Site Analogue with a Modified Binding-Site Nucleic Acid Base | | Descriptor: | DNA (30-MER), Poly(U)-binding-splicing factor PUF60 | | Authors: | Crichlow, G.V, Hsiao, H.-H, Albright, R, Lolis, E.J, Braddock, D.T. | | Deposit date: | 2016-07-15 | | Release date: | 2017-08-23 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | Unraveling the mechanism of recognition of the 3' splice site of the adenovirus major late promoter intron by the alternative splicing factor PUF60.
Plos One, 15, 2020
|
|
5W4F
 
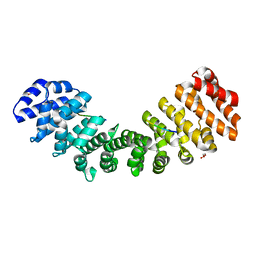 | | Importin binding to pol Mu NLS peptide | | Descriptor: | DNA-directed DNA/RNA polymerase mu, GLYCEROL, Importin subunit alpha-1 | | Authors: | Pedersen, L.C, London, R.E. | | Deposit date: | 2017-06-10 | | Release date: | 2018-06-13 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.984 Å) | | Cite: | Variations in nuclear localization strategies among pol X family enzymes.
Traffic, 2018
|
|
4LND
 
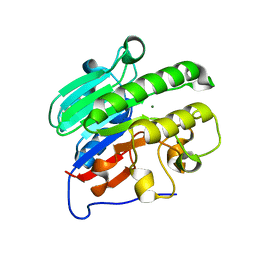 | | Crystal structure of human apurinic/apyrimidinic endonuclease 1 with essential Mg2+ cofactor | | Descriptor: | DNA-(apurinic or apyrimidinic site) lyase, MAGNESIUM ION | | Authors: | Manvilla, B.A, Pozharski, E, Toth, E.A, Drohat, A.C. | | Deposit date: | 2013-07-11 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure of human apurinic/apyrimidinic endonuclease 1 with the essential Mg(2+) cofactor.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
2KZD
 
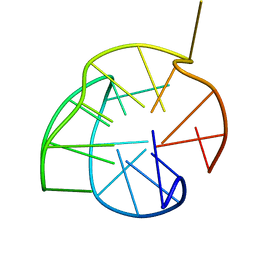 | | Structure of a (3+1) G-quadruplex formed by hTERT promoter sequence | | Descriptor: | DNA (5'-D(*AP*GP*GP*GP*IP*AP*GP*GP*GP*GP*CP*TP*GP*GP*GP*AP*GP*GP*GP*C)-3') | | Authors: | Lim, K.W, Lacroix, L, Yue, D.J.E, Lim, J.K.C, Lim, J.M.W, Phan, A.T. | | Deposit date: | 2010-06-16 | | Release date: | 2010-10-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Coexistence of two distinct G-quadruplex conformations in the hTERT promoter
J.Am.Chem.Soc., 132, 2010
|
|
2KZE
 
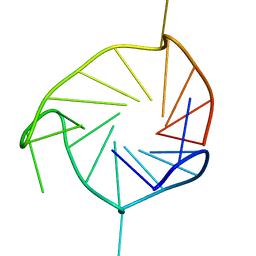 | | Structure of an all-parallel-stranded G-quadruplex formed by hTERT promoter sequence | | Descriptor: | DNA (5'-D(*AP*IP*GP*GP*GP*AP*GP*GP*GP*IP*CP*TP*GP*GP*GP*AP*GP*GP*GP*C)-3') | | Authors: | Lim, K.W, Lacroix, L, Yue, D.J.E, Lim, J.K.C, Lim, J.M.W, Phan, A.T. | | Deposit date: | 2010-06-16 | | Release date: | 2010-10-06 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Coexistence of two distinct G-quadruplex conformations in the hTERT promoter
J.Am.Chem.Soc., 132, 2010
|
|
3OS2
 
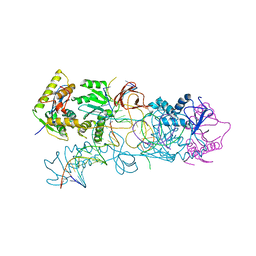 | | PFV target capture complex (TCC) at 3.32 A resolution | | Descriptor: | DNA (5'-D(*AP*TP*TP*GP*TP*CP*AP*TP*GP*GP*AP*AP*TP*TP*TP*CP*GP*CP*A)-3'), DNA (5'-D(*CP*CP*CP*GP*AP*GP*GP*CP*AP*CP*GP*TP*GP*CP*TP*AP*GP*CP*AP*CP*GP*TP*GP*CP*CP*TP*CP*GP*GP*G)-3'), DNA (5'-D(*TP*GP*CP*GP*AP*AP*AP*TP*TP*CP*CP*AP*TP*GP*AP*CP*A)-3'), ... | | Authors: | Maertens, G.N, Hare, S, Cherepanov, P. | | Deposit date: | 2010-09-08 | | Release date: | 2010-11-17 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | The mechanism of retroviral integration from X-ray structures of its key intermediates
Nature, 468, 2010
|
|
