8F2H
 
 | |
6TLJ
 
 | |
2Y4W
 
 | | Solution structure of human ubiquitin conjugating enzyme Rad6b | | Descriptor: | UBIQUITIN-CONJUGATING ENZYME E2 B | | Authors: | Huang, A, Hibbert, R.G, deJong, R.N, Das, D, Sixma, T.K, Boelens, R. | | Deposit date: | 2011-01-11 | | Release date: | 2011-05-11 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Symmetry and Asymmetry of the Ring-Ring Dimer of Rad18
J.Mol.Biol., 410, 2011
|
|
4Q5H
 
 | |
4Q5E
 
 | |
8ODR
 
 | | Mimetic of UBC9-SUMO1 | | Descriptor: | SULFATE ION, SUMO-conjugating enzyme UBC9, Small ubiquitin-related modifier 1 | | Authors: | Coste, F, Goffinont, S, Suskiewicz, M.J. | | Deposit date: | 2023-03-09 | | Release date: | 2023-06-07 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural insights into the regulation of the human E2∼SUMO conjugate through analysis of its stable mimetic.
J.Biol.Chem., 299, 2023
|
|
2GJD
 
 | | Distinct functional domains of Ubc9 dictate cell survival and resistance to genotoxic stress | | Descriptor: | Ubiquitin-conjugating enzyme E2-18 kDa | | Authors: | van Waardenburg, R.C, Duda, D.M, Lancaster, C.S, Schulman, B.A, Bjornsti, M.A. | | Deposit date: | 2006-03-30 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Distinct functional domains of ubc9 dictate cell survival and resistance to genotoxic stress.
Mol.Cell.Biol., 26, 2006
|
|
6LQS
 
 | |
2EWN
 
 | | Ecoli Biotin Repressor with co-repressor analog | | Descriptor: | ((2R,3S,4R,5R)-5-(6-AMINO-9H-PURIN-9-YL)-3,4-DIHYDROXY-TETRAHYDROFURAN-2-YL)METHYL 5-((3AS,4S,6AR)-2-OXO-HEXAHYDRO-1H-THIENO[3,4-D]IMIDAZOL-4-YL)PENTYL HYDROGEN PHOSPHATE, BirA bifunctional protein | | Authors: | Wood, Z.A, Weaver, L.H, Matthews, B.W. | | Deposit date: | 2005-11-04 | | Release date: | 2006-02-21 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Co-repressor Induced Order and Biotin Repressor Dimerization: A Case for Divergent Followed by Convergent Evolution.
J.Mol.Biol., 357, 2006
|
|
7BBF
 
 | |
4Y1L
 
 | | Ubc9 Homodimer The Missing Link in Poly-SUMO Chain Formation | | Descriptor: | RWD domain-containing protein 3, SUMO-conjugating enzyme UBC9 | | Authors: | Aileen, Y.A, Ambaye, N.D, Li, Y.J, Vega, R, Bzymek, K, Williams, J.C, Hu, W, Chen, Y. | | Deposit date: | 2015-02-08 | | Release date: | 2015-05-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | RWD Domain as an E2 (Ubc9)-Interaction Module.
J.Biol.Chem., 290, 2015
|
|
6NIF
 
 | | crystal structure of human REV7-RAN complex | | Descriptor: | hREV7, GTP-binding nuclear protein Ran, hREV3 fusion | | Authors: | Wang, X, Pertz, L, Hua, D.P, Zhang, T.Q, Listovsky, T, Xie, W. | | Deposit date: | 2018-12-27 | | Release date: | 2019-09-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.002 Å) | | Cite: | REV7 has a dynamic adaptor region to accommodate small GTPase RAN/ShigellaIpaB ligands, and its activity is regulated by the RanGTP/GDP switch.
J.Biol.Chem., 294, 2019
|
|
4DHZ
 
 | | The structure of h/ceOTUB1-ubiquitin aldehyde-UBC13~Ub | | Descriptor: | Ubiquitin, Ubiquitin aldehyde, Ubiquitin thioesterase otubain-like, ... | | Authors: | Wiener, R, Zhang, X, Wang, T, Wolberger, C. | | Deposit date: | 2012-01-30 | | Release date: | 2012-02-22 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | The mechanism of OTUB1-mediated inhibition of ubiquitination.
Nature, 483, 2012
|
|
3C6O
 
 | |
3C6P
 
 | |
6Y6X
 
 | | Tetracenomycin X bound to the human ribosome | | Descriptor: | 28S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Buschauer, R, Cheng, J, Berninghausen, O, Beckmann, R, Wilson, D.N. | | Deposit date: | 2020-02-27 | | Release date: | 2020-07-08 | | Last modified: | 2023-11-08 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Tetracenomycin X inhibits translation by binding within the ribosomal exit tunnel.
Nat.Chem.Biol., 16, 2020
|
|
3C6N
 
 | | Small molecule agonists and antagonists of F-box protein-substrate interactions in auxin perception and signaling | | Descriptor: | (2S)-8-[(tert-butoxycarbonyl)amino]-2-(1H-indol-3-yl)octanoic acid, INOSITOL HEXAKISPHOSPHATE, SKP1-like protein 1A, ... | | Authors: | Tan, X, Zheng, N, Hayashi, K. | | Deposit date: | 2008-02-04 | | Release date: | 2008-04-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Small-molecule agonists and antagonists of F-box protein-substrate interactions in auxin perception and signaling.
Proc.Natl.Acad.Sci.Usa, 105, 2008
|
|
4WID
 
 | | Crystal structure of the immediate-early 1 protein (IE1) at 2.31 angstrom (tetragonal form after crystal dehydration) | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, RhUL123 | | Authors: | Klingl, S, Scherer, M, Sevvana, M, Muller, Y.A, Stamminger, T. | | Deposit date: | 2014-09-25 | | Release date: | 2014-10-29 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Crystal Structure of Cytomegalovirus IE1 Protein Reveals Targeting of TRIM Family Member PML via Coiled-Coil Interactions.
Plos Pathog., 10, 2014
|
|
8GYM
 
 | | Cryo-EM structure of Tetrahymena thermophila respiratory mega-complex MC IV2+(I+III2+II)2 | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, 1,2-Distearoyl-sn-glycerophosphoethanolamine, ... | | Authors: | Wu, M.C, Hu, Y.Q, Han, F.Z, Zhou, L. | | Deposit date: | 2022-09-23 | | Release date: | 2023-06-14 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.96 Å) | | Cite: | Structures of Tetrahymena thermophila respiratory megacomplexes on the tubular mitochondrial cristae.
Nat Commun, 14, 2023
|
|
4YE8
 
 | |
4RQW
 
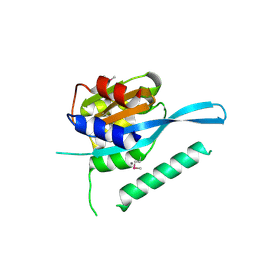 | | Crystal structure of Myc3 N-terminal JAZ-binding domain [44-238] from Arabidopsis | | Descriptor: | CALCIUM ION, Transcription factor MYC3 | | Authors: | Ke, J, Zhang, F, Zhou, X.E, Brunzelle, J, Zhou, M, Xu, H.E, Melcher, K, He, S.Y. | | Deposit date: | 2014-11-05 | | Release date: | 2015-08-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis of JAZ repression of MYC transcription factors in jasmonate signalling.
Nature, 525, 2015
|
|
4RRU
 
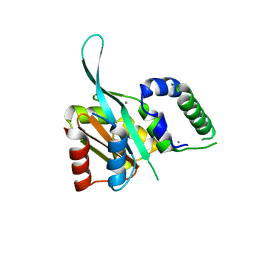 | | Myc3 N-terminal JAZ-binding domain[5-242] from arabidopsis | | Descriptor: | CALCIUM ION, Transcription factor MYC3 | | Authors: | Ke, J, Zhang, F, Zhou, X.E, Brunzelle, J.S, Zhou, M, Xu, H.E, Melcher, K, He, S.Y. | | Deposit date: | 2014-11-06 | | Release date: | 2015-08-12 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of JAZ repression of MYC transcription factors in jasmonate signalling.
Nature, 525, 2015
|
|
4RS9
 
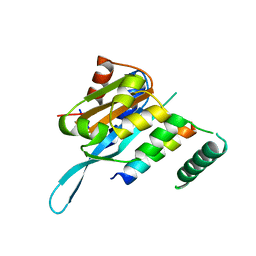 | | Structure of Myc3 N-terminal JAZ-binding domain [44-238] in complex with Jas motif of JAZ9 | | Descriptor: | Protein TIFY 7, Transcription factor MYC3 | | Authors: | Ke, J, Zhang, F, Zhou, X.E, Brunzelle, J.S, Zhou, M, Xu, H.E, Melcher, K, He, S.Y. | | Deposit date: | 2014-11-07 | | Release date: | 2015-08-12 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis of JAZ repression of MYC transcription factors in jasmonate signalling.
Nature, 525, 2015
|
|
4YE9
 
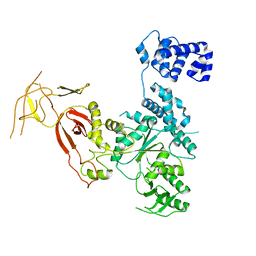 | |
4YE6
 
 | |
