4MTY
 
 | | Structure at 1A resolution of a helical aromatic foldamer-protein complex. | | Descriptor: | 4-(HYDROXYMERCURY)BENZOIC ACID, Carbonic anhydrase 2, GLYCEROL, ... | | Authors: | Ogayone, T, Buratto, J, Langlois D'Estaintot, B, Stupfel, M, Granier, T, Gallois, B, Huc, Y. | | Deposit date: | 2013-09-20 | | Release date: | 2013-12-11 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1 Å) | | Cite: | Structure of a complex formed by a protein and a helical aromatic oligoamide foldamer at 2.1 a resolution.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4MUC
 
 | | The 4th and 5th C-terminal domains of Factor H related protein 1 | | Descriptor: | Complement factor H-related protein 1, SULFATE ION | | Authors: | Bhattacharjee, A, Goldman, A, Kolodziejczyk, R, Jokiranta, T.S. | | Deposit date: | 2013-09-21 | | Release date: | 2015-02-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.897 Å) | | Cite: | The Major Autoantibody Epitope on Factor H in Atypical Hemolytic Uremic Syndrome Is Structurally Different from Its Homologous Site in Factor H-related Protein 1, Supporting a Novel Model for Induction of Autoimmunity in This Disease.
J.Biol.Chem., 290, 2015
|
|
2VWW
 
 | | ephB4 kinase domain inhibitor complex | | Descriptor: | EPHRIN TYPE-B RECEPTOR 4, N'-(5-CHLORO-1,3-BENZODIOXOL-4-YL)-N-(3,4,5- TRIMETHOXYPHENYL)PYRIMIDINE-2,4-DIAMINE | | Authors: | Read, J, Brassington, C.A, Green, I, McCall, E.J, Valentine, A.L, Barratt, D, Leach, A.G, Kettle, J.G. | | Deposit date: | 2008-06-27 | | Release date: | 2008-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Inhibitors of the Tyrosine Kinase Ephb4. Part 1: Structure-Based Design and Optimization of a Series of 2,4-Bis-Anilinopyrimidines
Bioorg.Med.Chem.Lett., 18, 2008
|
|
4MW4
 
 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with inhibitor 1-{3-[(5-chloro-2-hydroxy-3-iodobenzyl)amino]propyl}-3-thiophen-3-ylurea (Chem 1473) | | Descriptor: | 1-{3-[(5-chloro-2-hydroxy-3-iodobenzyl)amino]propyl}-3-thiophen-3-ylurea, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Koh, C.Y, Kim, J.E, Wetzel, A.B, de van der Schueren, W.J, Shibata, S, Liu, J, Zhang, Z, Fan, E, Verlinde, C.L.M.J, Hol, W.G.J. | | Deposit date: | 2013-09-24 | | Release date: | 2014-04-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structures of Trypanosoma brucei Methionyl-tRNA Synthetase with Urea-Based Inhibitors Provide Guidance for Drug Design against Sleeping Sickness.
Plos Negl Trop Dis, 8, 2014
|
|
3DN0
 
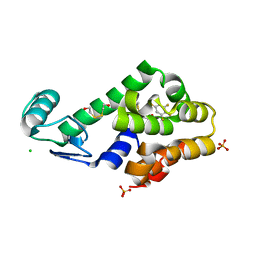 | | Pentafluorobenzene binding in the hydrophobic cavity of T4 lysozyme L99A mutant | | Descriptor: | 1,2,3,4,5-pentafluorobenzene, 2-HYDROXYETHYL DISULFIDE, CHLORIDE ION, ... | | Authors: | Liu, L, Matthews, B.W. | | Deposit date: | 2008-07-01 | | Release date: | 2008-11-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Halogenated benzenes bound within a non-polar cavity in T4 lysozyme provide examples of I...S and I...Se halogen-bonding.
J.Mol.Biol., 385, 2009
|
|
4MWB
 
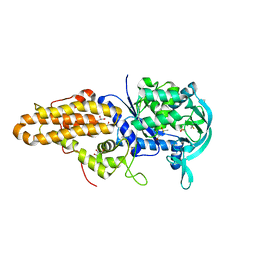 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with inhibitor 1-(3-{[(2,5-dichlorothiophen-3-yl)methyl]amino}propyl)-3-thiophen-3-ylurea (Chem 1509) | | Descriptor: | 1-(3-{[(2,5-dichlorothiophen-3-yl)methyl]amino}propyl)-3-thiophen-3-ylurea, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Koh, C.Y, Kim, J.E, Wetzel, A.B, de van der Schueren, W.J, Shibata, S, Liu, J, Zhang, Z, Fan, E, Verlinde, C.L.M.J, Hol, W.G.J. | | Deposit date: | 2013-09-24 | | Release date: | 2014-04-30 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.313 Å) | | Cite: | Structures of Trypanosoma brucei Methionyl-tRNA Synthetase with Urea-Based Inhibitors Provide Guidance for Drug Design against Sleeping Sickness.
Plos Negl Trop Dis, 8, 2014
|
|
1LTM
 
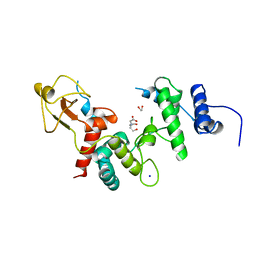 | | ACCELERATED X-RAY STRUCTURE ELUCIDATION OF A 36 KDA MURAMIDASE/TRANSGLYCOSYLASE USING WARP | | Descriptor: | 1,2-ETHANEDIOL, 36 KDA SOLUBLE LYTIC TRANSGLYCOSYLASE, BICINE, ... | | Authors: | Van Asselt, E.J, Dijkstra, B.W. | | Deposit date: | 1997-09-26 | | Release date: | 1998-11-11 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Accelerated X-ray structure elucidation of a 36 kDa muramidase/transglycosylase using wARP.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
5C8Z
 
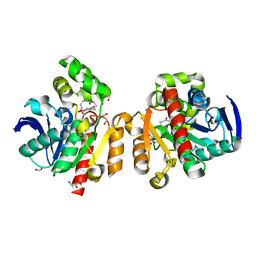 | | ZHD-ZGR complex after ZHD crystal soaking in ZEN for 30min | | Descriptor: | 2,4-dihydroxy-6-[(1E,10S)-10-hydroxy-6-oxoundec-1-en-1-yl]benzoic acid, FORMIC ACID, GLYCEROL, ... | | Authors: | Hu, X.-J, Qi, Q, Yang, W.-J. | | Deposit date: | 2015-06-26 | | Release date: | 2016-06-29 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The structure of a complex of the lactonohydrolase zearalenone hydrolase with the hydrolysis product of zearalenone at 1.60 angstrom resolution.
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
5AZG
 
 | | Crystal structure of LGG-1 complexed with a UNC-51 peptide | | Descriptor: | CADMIUM ION, Protein lgg-1, Serine/threonine-protein kinase unc-51 | | Authors: | Watanabe, Y, Fujioka, Y, Noda, N.N. | | Deposit date: | 2015-10-05 | | Release date: | 2015-12-30 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Structural Basis of the Differential Function of the Two C. elegans Atg8 Homologs, LGG-1 and LGG-2, in Autophagy.
Mol.Cell, 60, 2015
|
|
7FZV
 
 | | Crystal Structure of human FABP4 in complex with 1-[(2,4-dichlorophenyl)methyl]-4-hydroxy-3-[(E)-3-phenylprop-2-enyl]pyridin-2-one, i.e. SMILES C1(=C(C=CN(C1=O)Cc1c(cc(cc1)Cl)Cl)O)C/C=C/c1ccccc1 with IC50=0.557 microM | | Descriptor: | 1-[(2,4-dichlorophenyl)methyl]-4-hydroxy-3-[(2E)-3-phenylprop-2-en-1-yl]pyridin-2(1H)-one, Fatty acid-binding protein, adipocyte, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Brunner, M, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
3SRE
 
 | | Serum paraoxonase-1 by directed evolution at pH 6.5 | | Descriptor: | BROMIDE ION, CALCIUM ION, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Ben David, M, Elias, M, Silman, I, Sussman, J.L, Tawfik, D.S. | | Deposit date: | 2011-07-07 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Catalytic versatility and backups in enzyme active sites: the case of serum paraoxonase 1.
J.Mol.Biol., 418, 2012
|
|
2VWU
 
 | | ephB4 kinase domain inhibitor complex | | Descriptor: | EPHRIN TYPE-B RECEPTOR 4, N-(5-chloro-1,3-benzodioxol-4-yl)-6-methoxy-7-(3-piperidin-1-ylpropoxy)quinazolin-4-amine | | Authors: | Read, J, Brassington, C.A, Green, I, McCall, E.J, Valentine, A.L, Barratt, D, Rowsell, S, Packer, M, McAlister, M. | | Deposit date: | 2008-06-27 | | Release date: | 2008-07-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Inhibitors of the Tyrosine Kinase Ephb4. Part 1: Structure-Based Design and Optimization of a Series of 2,4-Bis-Anilinopyrimidines
Bioorg.Med.Chem.Lett., 18, 2008
|
|
7G17
 
 | | Crystal Structure of human FABP4 in complex with 2-(1H-indol-7-yloxymethyl)-1,3-thiazole-4-carboxylic acid | | Descriptor: | 1,2-ETHANEDIOL, 2-{[(1H-indol-7-yl)oxy]methyl}-1,3-thiazole-4-carboxylic acid, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Brunner, M, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
4MVX
 
 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with inhibitor 1-{3-[(3,5-dichlorobenzyl)amino]propyl}-3-phenylurea (Chem 1356) | | Descriptor: | 1-{3-[(3,5-dichlorobenzyl)amino]propyl}-3-phenylurea, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Koh, C.Y, Kim, J.E, Wetzel, A.B, de van der Schueren, W.J, Shibata, S, Liu, J, Zhang, Z, Fan, E, Verlinde, C.L.M.J, Hol, W.G.J. | | Deposit date: | 2013-09-24 | | Release date: | 2014-04-30 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structures of Trypanosoma brucei Methionyl-tRNA Synthetase with Urea-Based Inhibitors Provide Guidance for Drug Design against Sleeping Sickness.
Plos Negl Trop Dis, 8, 2014
|
|
1PN2
 
 | | Crystal structure analysis of the selenomethionine labelled 2-enoyl-CoA hydratase 2 domain of Candida tropicalis multifunctional enzyme type 2 | | Descriptor: | 1,2-ETHANEDIOL, Peroxisomal hydratase-dehydrogenase-epimerase | | Authors: | Koski, M.K, Haapalainen, A.M, Hiltunen, J.K, Glumoff, T. | | Deposit date: | 2003-06-12 | | Release date: | 2004-04-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | A Two-domain Structure of One Subunit Explains Unique Features of Eukaryotic Hydratase 2.
J.Biol.Chem., 279, 2004
|
|
6GDO
 
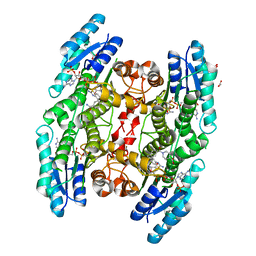 | | Trypanosoma brucei PTR1 in complex with inhibitor 2g (F240) | | Descriptor: | ACETATE ION, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Pozzi, C, Landi, G, Mangani, S. | | Deposit date: | 2018-04-24 | | Release date: | 2019-04-03 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Enhancement of Benzothiazoles as Pteridine Reductase-1 Inhibitors for the Treatment of Trypanosomatidic Infections.
J.Med.Chem., 62, 2019
|
|
4MW9
 
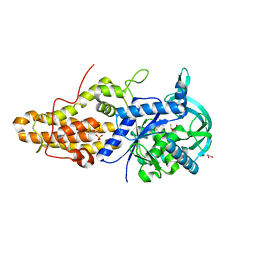 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with inhibitor 1-{3-[(3-ethynylbenzyl)amino]propyl}-3-thiophen-3-ylurea (Chem 1478) | | Descriptor: | 1-{3-[(3-ethynylbenzyl)amino]propyl}-3-thiophen-3-ylurea, DIMETHYL SULFOXIDE, GLYCEROL, ... | | Authors: | Koh, C.Y, Kim, J.E, Wetzel, A.B, de van der Schueren, W.J, Shibata, S, Liu, J, Zhang, Z, Fan, E, Verlinde, C.L.M.J, Hol, W.G.J. | | Deposit date: | 2013-09-24 | | Release date: | 2014-04-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structures of Trypanosoma brucei Methionyl-tRNA Synthetase with Urea-Based Inhibitors Provide Guidance for Drug Design against Sleeping Sickness.
Plos Negl Trop Dis, 8, 2014
|
|
5B2Q
 
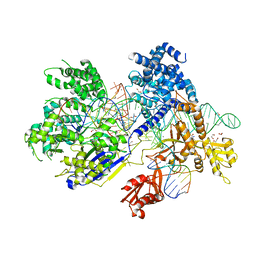 | | Crystal structure of Francisella novicida Cas9 RHA in complex with sgRNA and target DNA (TGG PAM) | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CALCIUM ION, ... | | Authors: | Hirano, H, Nishimasu, H, Nakane, T, Ishitani, R, Nureki, O. | | Deposit date: | 2016-02-01 | | Release date: | 2016-03-02 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure and Engineering of Francisella novicida Cas9
Cell, 164, 2016
|
|
4TQN
 
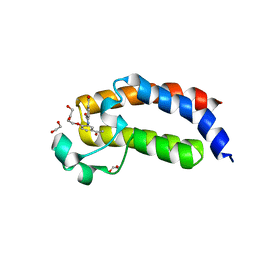 | |
7FZU
 
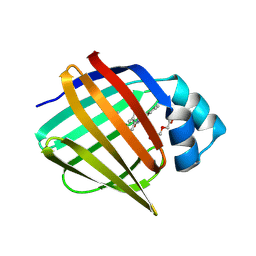 | | Crystal Structure of human FABP4 binding site mutated to that of FABP5 in complex with 2-(indole-1-carbonylamino)benzoic acid, i.e. SMILES c12N(C(=O)Nc3c(cccc3)C(=O)O)C=Cc1cccc2 with IC50=18.1696 microM | | Descriptor: | 1,2-ETHANEDIOL, 2-[(2,3-dihydro-1H-indole-1-carbonyl)amino]benzoic acid, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Ning, R, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
1V5V
 
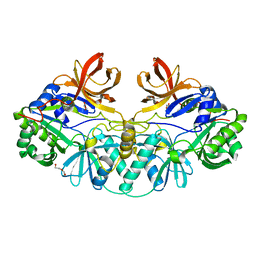 | |
6UMT
 
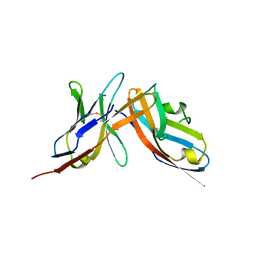 | | High-affinity human PD-1 PD-L2 complex | | Descriptor: | MAGNESIUM ION, Programmed cell death 1 ligand 2, Programmed cell death protein 1 | | Authors: | Tang, S, Kim, P.S. | | Deposit date: | 2019-10-10 | | Release date: | 2019-11-27 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.986 Å) | | Cite: | A high-affinity human PD-1/PD-L2 complex informs avenues for small-molecule immune checkpoint drug discovery.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
3EUY
 
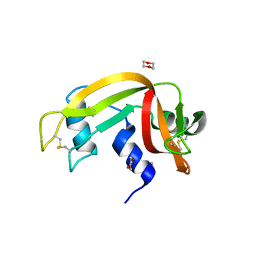 | | Crystal Structure of Ribonuclease A in 50% Dioxane | | Descriptor: | 1,4-DIETHYLENE DIOXIDE, Ribonuclease pancreatic | | Authors: | Dechene, M, Wink, G, Smith, M, Swartz, P, Mattos, C. | | Deposit date: | 2008-10-12 | | Release date: | 2009-06-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Multiple solvent crystal structures of ribonuclease A: An assessment of the method
Proteins, 76, 2009
|
|
5VJP
 
 | | Crystal Structure of Adenine Phosphoribosyltransferase from Saccharomyces cerevisiae Complexed with L-2,5-Dideoxy-2,5-Imino-Altritol 1,6-Bisphosphate (L-DIAB) and Adenine | | Descriptor: | 1,2-ETHANEDIOL, ADENINE, Adenine phosphoribosyltransferase 1, ... | | Authors: | Harijan, R.K, Ducati, R.G, Bonanno, J.B, Almo, S.C, Schramm, V.L. | | Deposit date: | 2017-04-19 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Synthesis of bis-Phosphate Iminoaltritol Enantiomers and Structural Characterization with Adenine Phosphoribosyltransferase.
ACS Chem. Biol., 13, 2018
|
|
7FYG
 
 | | Crystal Structure of human FABP4 in complex with 6-benzyl-2H-1,2,4-triazine-3,5-dithione | | Descriptor: | 1,2-ETHANEDIOL, 6-benzyl-1,2,4-triazine-3,5(2H,4H)-dithione, Fatty acid-binding protein, ... | | Authors: | Ehler, A, Benz, J, Obst, U, Koch, W, Rudolph, M.G. | | Deposit date: | 2023-04-27 | | Release date: | 2023-06-14 | | Last modified: | 2025-08-13 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | A high-resolution data set of fatty acid-binding protein structures. III. Unexpectedly high occurrence of wrong ligands.
Acta Crystallogr D Struct Biol, 81, 2025
|
|
