5J0W
 
 | |
3GDX
 
 | | Dna polymerase beta with a gapped DND substrate and dTMP(CF2)PP | | Descriptor: | 5'-D(*CP*CP*GP*AP*CP*AP*GP*CP*GP*CP*AP*TP*CP*AP*GP*C)-3', 5'-D(*GP*CP*TP*GP*AP*TP*GP*CP*GP*C)-3', 5'-D(P*GP*TP*CP*GP*G)-3', ... | | Authors: | Wilson, S.H, Batra, V.K, Pedersen, L.C. | | Deposit date: | 2009-02-24 | | Release date: | 2009-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Alpha,beta-difluoromethylene deoxynucleoside 5'-triphosphates: a convenient synthesis of useful probes for DNA polymerase beta structure and function
Org.Lett., 11, 2009
|
|
5J0Y
 
 | |
5J0P
 
 | |
5J0S
 
 | |
5J0Q
 
 | |
5J0T
 
 | |
5J0U
 
 | |
8V55
 
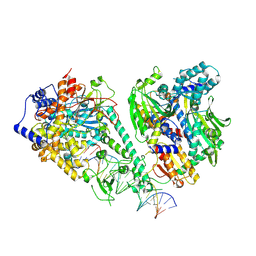 | | Human mitochondrial DNA polymerase gamma bound to a replication fork in an open conformation | | Descriptor: | DNA polymerase subunit gamma-1, DNA polymerase subunit gamma-2, mitochondrial, ... | | Authors: | Riccio, A.A, Krahn, J.M, Bouvette, J, Borgnia, M.J, Copeland, W.C. | | Deposit date: | 2023-11-30 | | Release date: | 2024-07-10 | | Last modified: | 2024-07-31 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Coordinated DNA polymerization by Pol gamma and the region of LonP1 regulated proteolysis.
Nucleic Acids Res., 52, 2024
|
|
8V54
 
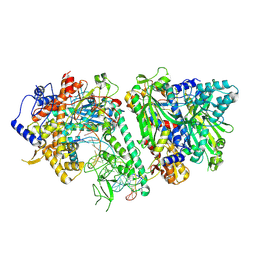 | | Engaged conformation of the human mitochondrial DNA polymerase gamma bound to DNA | | Descriptor: | DNA (25-MER), DNA (44-MER), DNA polymerase subunit gamma-1, ... | | Authors: | Riccio, A.A, Krahn, J.M, Bouvette, J, Borgnia, J.M, Copeland, W.C. | | Deposit date: | 2023-11-30 | | Release date: | 2024-07-10 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Coordinated DNA polymerization by Pol gamma and the region of LonP1 regulated proteolysis.
Nucleic Acids Res., 52, 2024
|
|
9FJR
 
 | |
8OJ6
 
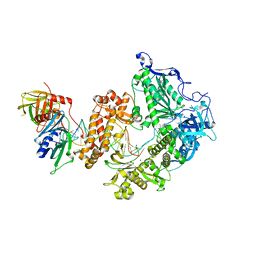 | | HSV-1 DNA polymerase-processivity factor complex in pre-translocation state | | Descriptor: | DNA (22-MER), DNA (48-MER), DNA polymerase catalytic subunit, ... | | Authors: | Gustavsson, E, Grunewald, K, Elias, P, Hallberg, B.M. | | Deposit date: | 2023-03-24 | | Release date: | 2024-04-03 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (2.41 Å) | | Cite: | Dynamics of the Herpes simplex virus DNA polymerase holoenzyme during DNA synthesis and proof-reading revealed by Cryo-EM.
Nucleic Acids Res., 52, 2024
|
|
4JL3
 
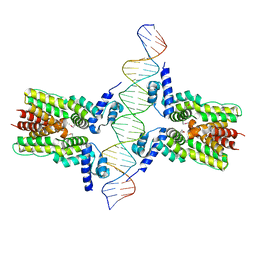 | | Crystal structure of ms6564-dna complex | | Descriptor: | DNA (31-MER), Transcriptional regulator, TetR family | | Authors: | Yang, S.F, Gao, Z.Q, He, Z.G, Dong, Y.H. | | Deposit date: | 2013-03-12 | | Release date: | 2013-06-26 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis for interaction between Mycobacterium smegmatis Ms6564, a TetR family master regulator, and its target DNA.
J.Biol.Chem., 288, 2013
|
|
1GLU
 
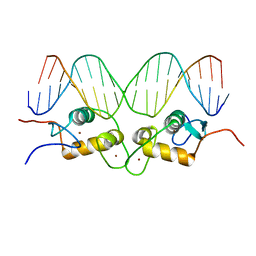 | | CRYSTALLOGRAPHIC ANALYSIS OF THE INTERACTION OF THE GLUCOCORTICOID RECEPTOR WITH DNA | | Descriptor: | DNA (5'-D(*CP*CP*AP*GP*AP*AP*CP*AP*TP*CP*GP*AP*TP*GP*TP*TP*C P*TP*G)-3'), PROTEIN (GLUCOCORTICOID RECEPTOR), ZINC ION | | Authors: | Luisi, B.F, Xu, W.X, Otwinowski, Z, Freedman, L.P, Yamamoto, K.R, Sigler, P.B. | | Deposit date: | 1992-08-30 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystallographic analysis of the interaction of the glucocorticoid receptor with DNA.
Nature, 352, 1991
|
|
5DB9
 
 | |
2KEK
 
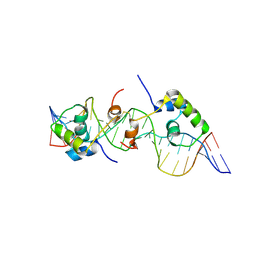 | | Solution structure of a dimer of LAC repressor DNA-binding domain complexed to its natural operator O3 | | Descriptor: | DNA (5'-D(*CP*GP*GP*CP*AP*GP*TP*GP*AP*GP*CP*GP*CP*AP*AP*CP*GP*CP*AP*AP*TP*TP*C)-3'), DNA (5'-D(P*GP*AP*AP*TP*TP*GP*CP*GP*TP*TP*GP*CP*GP*CP*TP*CP*AP*CP*TP*GP*CP*CP*G)-3'), Lactose operon repressor | | Authors: | Romanuka, J, Folkers, G, Biris, N, Tishchenko, E, Wienk, H, Kaptein, R, Boelens, R. | | Deposit date: | 2009-01-30 | | Release date: | 2009-05-19 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Specificity and affinity of Lac repressor for the auxiliary operators O2 and O3 are explained by the structures of their protein-DNA complexes.
J.Mol.Biol., 390, 2009
|
|
2KEI
 
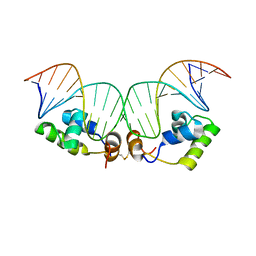 | | Refined Solution Structure of a Dimer of LAC repressor DNA-Binding domain complexed to its natural operator O1 | | Descriptor: | DNA (5'-D(*GP*AP*AP*TP*TP*GP*TP*GP*AP*GP*CP*GP*GP*AP*TP*AP*AP*CP*AP*AP*TP*TP*T)-3'), DNA (5'-D(P*AP*AP*AP*TP*TP*GP*TP*TP*AP*TP*CP*CP*GP*CP*TP*CP*AP*CP*AP*AP*TP*TP*C)-3'), Lactose operon repressor | | Authors: | Romanuka, J, Folkers, G, Biris, N, Tishchenko, E, Wienk, H, Kaptein, R, Boelens, R. | | Deposit date: | 2009-01-30 | | Release date: | 2009-05-19 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Specificity and affinity of Lac repressor for the auxiliary operators O2 and O3 are explained by the structures of their protein-DNA complexes.
J.Mol.Biol., 390, 2009
|
|
1HF0
 
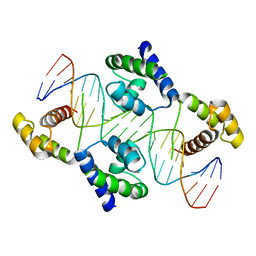 | | Crystal structure of the DNA-binding domain of Oct-1 bound to DNA as a dimer | | Descriptor: | DNA 5'-D(*CP*AP*CP*AP*TP*TP*TP*GP*AP*AP*AP*GP*GP* CP*AP*AP*AP*TP*GP*GP*AP*G)-3', DNA 5'-D(*CP*TP*CP*CP*AP*TP*TP*TP*GP*CP*CP*TP*TP* TP*CP*AP*AP*AP*TP*GP*TP*G)-3', OCTAMER-BINDING TRANSCRIPTION FACTOR 1 | | Authors: | Remenyi, A, Tomilin, A, Pohl, E, Scholer, H.R, Wilmanns, M. | | Deposit date: | 2000-11-27 | | Release date: | 2001-11-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Differential Dimer Activities of the Transcription Factor Oct-1 by DNA-Induced Interface Swapping
Mol.Cell, 8, 2001
|
|
5VXN
 
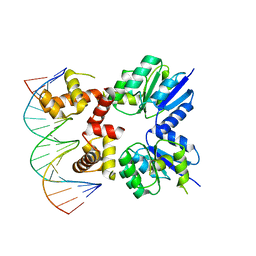 | | Structure of two RcsB dimers bound to two parallel DNAs. | | Descriptor: | DNA (5'-D(*GP*AP*TP*TP*TP*AP*GP*GP*AP*AP*AP*AP*AP*TP*CP*TP*TP*AP*GP*AP*TP*A)-3'), DNA (5'-D(*TP*TP*TP*AP*GP*GP*AP*AP*AP*AP*AP*TP*CP*TP*TP*AP*GP*A)-3'), Transcriptional regulatory protein RcsB | | Authors: | Filippova, E.V, Minasov, G, Pshenychnyi, S, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-05-23 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (3.375 Å) | | Cite: | Structural Basis for DNA Recognition by the Two-Component Response Regulator RcsB.
MBio, 9, 2018
|
|
4X0Q
 
 | |
2IRF
 
 | | CRYSTAL STRUCTURE OF AN IRF-2/DNA COMPLEX. | | Descriptor: | DNA (5'-D(*TP*TP*CP*AP*CP*TP*TP*TP*CP*AP*CP*(5IU)P*T)-3'), DNA (5'-D(P*AP*AP*GP*TP*GP*AP*AP*AP*GP*(5IU)P*GP*A)-3'), INTERFERON REGULATORY FACTOR 2, ... | | Authors: | Fujii, Y, Shimizu, T, Kusumoto, M, Kyogoku, Y, Taniguchi, T, Hakoshima, T. | | Deposit date: | 1999-05-30 | | Release date: | 1999-10-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of an IRF-DNA complex reveals novel DNA recognition and cooperative binding to a tandem repeat of core sequences.
EMBO J., 18, 1999
|
|
5DBB
 
 | |
4X0P
 
 | |
2KEJ
 
 | | Solution structure of a dimer of LAC repressor DNA-binding domain complexed to its natural operator O2 | | Descriptor: | DNA (5'-D(*GP*AP*AP*AP*TP*GP*TP*GP*AP*GP*CP*GP*AP*GP*TP*AP*AP*CP*AP*AP*CP*CP*G)-3'), DNA (5'-D(P*CP*GP*GP*TP*TP*GP*TP*TP*AP*CP*TP*CP*GP*CP*TP*CP*AP*CP*AP*TP*TP*TP*C)-3'), Lactose operon repressor | | Authors: | Romanuka, J, Folkers, G, Biris, N, Tishchenko, E, Wienk, H, Kaptein, R, Boelens, R. | | Deposit date: | 2009-01-30 | | Release date: | 2009-05-19 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Specificity and affinity of Lac repressor for the auxiliary operators O2 and O3 are explained by the structures of their protein-DNA complexes.
J.Mol.Biol., 390, 2009
|
|
1JEY
 
 | | Crystal Structure of the Ku heterodimer bound to DNA | | Descriptor: | DNA (34-MER), DNA (5'-D(*GP*TP*TP*TP*TP*TP*AP*GP*TP*TP*TP*AP*TP*TP*GP*GP*GP*CP*GP*CP*G)-3'), Ku70, ... | | Authors: | Walker, J.R, Corpina, R.A, Goldberg, J. | | Deposit date: | 2001-06-19 | | Release date: | 2001-08-10 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Ku heterodimer bound to DNA and its implications for double-strand break repair.
Nature, 412, 2001
|
|
