6L72
 
 | | Sirtuin 2 demyristoylation native final product | | Descriptor: | NAD-dependent protein deacetylase sirtuin-2, ZINC ION, [(2S,3R,4R,5R)-5-[[[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxymethyl]-2,4-bis(oxidanyl)oxolan-3-yl] tetradecanoate | | Authors: | Chen, L.F. | | Deposit date: | 2019-10-30 | | Release date: | 2021-03-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.501 Å) | | Cite: | Sirtuin 2 protein with H3K18 myristoylated peptide
To Be Published
|
|
6L71
 
 | |
4ZZH
 
 | | SIRT1/Activator Complex | | Descriptor: | (4S)-N-[3-(1,3-oxazol-5-yl)phenyl]-7-[3-(trifluoromethyl)phenyl]-3,4-dihydro-1,4-methanopyrido[2,3-b][1,4]diazepine-5(2H)-carboxamide, NAD-dependent protein deacetylase sirtuin-1, ZINC ION | | Authors: | Dai, H. | | Deposit date: | 2015-05-22 | | Release date: | 2015-07-15 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (3.1001 Å) | | Cite: | Crystallographic structure of a small molecule SIRT1 activator-enzyme complex.
Nat Commun, 6, 2015
|
|
4Y6L
 
 | | Human SIRT2 in complex with myristoylated peptide (H3K9myr) | | Descriptor: | NAD-dependent protein deacetylase sirtuin-2, ZINC ION, peptide THR-ALA-ARG-MYK-SER-THR-GLY | | Authors: | Kudo, N, Ito, A, Yoshida, M. | | Deposit date: | 2015-02-13 | | Release date: | 2016-01-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Kinetic and Structural Basis for Acyl-Group Selectivity and NAD(+) Dependence in Sirtuin-Catalyzed Deacylation.
Biochemistry, 54, 2015
|
|
3JWP
 
 | | Crystal structure of Plasmodium falciparum SIR2A (PF13_0152) in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, TRIETHYLENE GLYCOL, Transcriptional regulatory protein sir2 homologue, ... | | Authors: | Wernimont, A.K, Hutchinson, A, Lin, Y.H, MacKenzie, F, Senisterra, G, Allali-Hassanali, A, Vedadi, M, Ravichandran, M, Cossar, D, Kozieradzki, I, Zhao, Y, Schapira, M, Arrowsmith, C.H, Bountra, C, Weigelt, J, Edwards, A.M, Hui, R, Qiu, W, Brand, V, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-09-18 | | Release date: | 2009-10-20 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Crystal structure of Plasmodium falciparum SIR2A (PF13_0152) in complex with AMP
TO BE PUBLISHED
|
|
4ZZJ
 
 | | SIRT1/Activator/Substrate Complex | | Descriptor: | (3S)-1,3-dimethyl-N-[3-(1,3-oxazol-5-yl)phenyl]-6-[3-(trifluoromethyl)phenyl]-2,3-dihydropyrido[2,3-b]pyrazine-4(1H)-carboxamide, Ac-p53, CARBA-NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Dai, H. | | Deposit date: | 2015-05-22 | | Release date: | 2015-07-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.7403 Å) | | Cite: | Crystallographic structure of a small molecule SIRT1 activator-enzyme complex.
Nat Commun, 6, 2015
|
|
3JR3
 
 | | Sir2 bound to acetylated peptide | | Descriptor: | Acetylated Peptide, NAD-dependent deacetylase, ZINC ION | | Authors: | Hawse, W.F, Wolberger, C. | | Deposit date: | 2009-09-08 | | Release date: | 2009-09-29 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structure-based mechanism of ADP-ribosylation by sirtuins.
J.Biol.Chem., 284, 2009
|
|
4ZZI
 
 | | SIRT1/Activator/Inhibitor Complex | | Descriptor: | (3S)-1,3-dimethyl-N-[3-(1,3-oxazol-5-yl)phenyl]-6-[3-(trifluoromethyl)phenyl]-2,3-dihydropyrido[2,3-b]pyrazine-4(1H)-carboxamide, 4-(4-{2-[(methylsulfonyl)amino]ethyl}piperidin-1-yl)thieno[3,2-d]pyrimidine-6-carboxamide, NAD-dependent protein deacetylase sirtuin-1, ... | | Authors: | Dai, H. | | Deposit date: | 2015-05-22 | | Release date: | 2015-07-15 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.7346 Å) | | Cite: | Crystallographic structure of a small molecule SIRT1 activator-enzyme complex.
Nat Commun, 6, 2015
|
|
3K35
 
 | | Crystal Structure of Human SIRT6 | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, NAD-dependent deacetylase sirtuin-6, SULFATE ION, ... | | Authors: | Pan, P.W, Dong, A, Qiu, W, Loppnau, P, Wang, J, Ravichandran, M, Bochkarev, A, Bountra, C, Weigelt, J, Arrowsmith, C.H, Min, J, Edwards, A.M, Structural Genomics Consortium (SGC) | | Deposit date: | 2009-10-01 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure and biochemical functions of SIRT6.
J.Biol.Chem., 286, 2011
|
|
5BTR
 
 | | Crystal structure of SIRT1 in complex with resveratrol and an AMC-containing peptide | | Descriptor: | AMC-containing peptide, NAD-dependent protein deacetylase sirtuin-1, RESVERATROL, ... | | Authors: | Cao, D, Wang, M, Qiu, X, Liu, D, Jiang, H, Yang, N, Xu, R.M. | | Deposit date: | 2015-06-03 | | Release date: | 2015-07-08 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Structural basis for allosteric, substrate-dependent stimulation of SIRT1 activity by resveratrol
Genes Dev., 29, 2015
|
|
1J8F
 
 | |
1ICI
 
 | | CRYSTAL STRUCTURE OF A SIR2 HOMOLOG-NAD COMPLEX | | Descriptor: | NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRANSCRIPTIONAL REGULATORY PROTEIN, SIR2 FAMILY, ... | | Authors: | Min, J, Landry, J, Sternglanz, R, Xu, R.-M. | | Deposit date: | 2001-04-01 | | Release date: | 2001-05-02 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of a SIR2 homolog-NAD complex.
Cell(Cambridge,Mass.), 105, 2001
|
|
1M2K
 
 | | Sir2 homologue F159A mutant-ADP ribose complex | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Silent Information Regulator 2, ZINC ION | | Authors: | Chang, J, Cho, Y. | | Deposit date: | 2002-06-24 | | Release date: | 2003-04-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural basis for the NAD-dependent deacetylase mechanism of Sir2
J.BIOL.CHEM., 277, 2002
|
|
1M2G
 
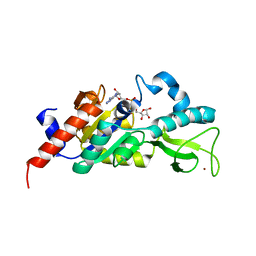 | | Sir2 homologue-ADP ribose complex | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Silent Information Regulator 2, ZINC ION | | Authors: | Chang, J, Cho, Y. | | Deposit date: | 2002-06-24 | | Release date: | 2003-04-08 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for the NAD-dependent deacetylase mechanism of Sir2
J.BIOL.CHEM., 277, 2002
|
|
1M2J
 
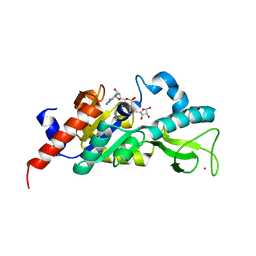 | | Sir2 homologue H80N mutant-ADP ribose complex | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Silent Information Regulator 2, ZINC ION | | Authors: | Chang, J, Cho, Y. | | Deposit date: | 2002-06-24 | | Release date: | 2003-04-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for the NAD-dependent deacetylase mechanism of Sir2
J.BIOL.CHEM., 277, 2002
|
|
1MA3
 
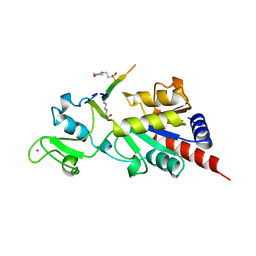 | | Structure of a Sir2 enzyme bound to an acetylated p53 peptide | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Cellular tumor antigen p53, Transcriptional regulatory protein, ... | | Authors: | Avalos, J.L, Celic, I, Muhammad, S, Cosgrove, M.S, Boeke, J.D, Wolberger, C. | | Deposit date: | 2002-07-31 | | Release date: | 2002-10-16 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure of a Sir2 enzyme bound to an acetylated p53 peptide
Mol.Cell, 10, 2002
|
|
1M2N
 
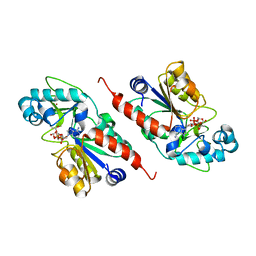 | |
1M2H
 
 | | Sir2 homologue S24A mutant-ADP ribose complex | | Descriptor: | ADENOSINE-5-DIPHOSPHORIBOSE, Silent Information Regulator 2, ZINC ION | | Authors: | Chang, J, Cho, Y. | | Deposit date: | 2002-06-24 | | Release date: | 2003-04-08 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for the NAD-dependent deacetylase mechanism of Sir2
J.BIOL.CHEM., 277, 2002
|
|
6RXQ
 
 | | Crystal structure of CobB Ac2 (A76G,I131C,V162A) in complex with H4K16Cr-2'OH-ADPr peptide intermediate after soaking | | Descriptor: | Histone H4, NAD-dependent protein deacylase, [[(2~{R},3~{S},4~{R},5~{R})-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl] [(2~{R},3~{R},4~{R},5~{S})-4-[(~{E})-but-2-enoxy]-3,5-bis(oxidanyl)oxolan-2-yl]methyl hydrogen phosphate | | Authors: | Spinck, M, Gasper, R, Neumann, H. | | Deposit date: | 2019-06-08 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Evolved, Selective Erasers of Distinct Lysine Acylations.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6RXK
 
 | | Crystal structure of CobB wt in complex with H4K16-Butyryl peptide | | Descriptor: | Histone H4, NAD-dependent protein deacylase, ZINC ION | | Authors: | Spinck, M, Gasper, R, Neumann, H. | | Deposit date: | 2019-06-08 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Evolved, Selective Erasers of Distinct Lysine Acylations.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
6RXS
 
 | | Crystal structure of CobB Ac3(A76G,Y92A, I131L, V187Y) in complex with H4K16-Acetyl peptide | | Descriptor: | GLYCEROL, Histone H4, NAD-dependent protein deacylase, ... | | Authors: | Spinck, M, Gasper, R, Neumann, H. | | Deposit date: | 2019-06-08 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.599 Å) | | Cite: | Evolved, Selective Erasers of Distinct Lysine Acylations.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
5BWN
 
 | | Crystal Structure of SIRT3 with a H3K9 Peptide Containing a Myristoyl Lysine | | Descriptor: | NAD-dependent protein deacetylase sirtuin-3, mitochondrial, ZINC ION, ... | | Authors: | Gai, W, Liu, D. | | Deposit date: | 2015-06-08 | | Release date: | 2016-07-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.942 Å) | | Cite: | Crystal structures of SIRT3 reveal that the alpha 2-alpha 3 loop and alpha 3-helix affect the interaction with long-chain acyl lysine.
Febs Lett., 590, 2016
|
|
4FZ3
 
 | | Crystal structure of SIRT3 in complex with acetyl p53 peptide coupled with 4-amino-7-methylcoumarin | | Descriptor: | NAD-dependent protein deacetylase sirtuin-3, mitochondrial, ZINC ION, ... | | Authors: | Liu, D, Wu, J, Zhang, D, Chen, K, Jiang, H, Liu, H. | | Deposit date: | 2012-07-06 | | Release date: | 2013-03-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery and Mechanism Study of SIRT1 Activators that Promote the Deacetylation of Fluorophore-Labeled Substrate
J.Med.Chem., 56, 2013
|
|
5BWO
 
 | | Crystal Structure of Human SIRT3 in Complex with a Palmitoyl H3K9 Peptide | | Descriptor: | NAD-dependent protein deacetylase sirtuin-3, mitochondrial, PALMITIC ACID, ... | | Authors: | Gai, W, Jiang, H, Liu, D. | | Deposit date: | 2015-06-08 | | Release date: | 2016-07-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.376 Å) | | Cite: | Crystal structures of SIRT3 reveal that the alpha 2-alpha 3 loop and alpha 3-helix affect the interaction with long-chain acyl lysine.
Febs Lett., 590, 2016
|
|
6RXJ
 
 | | Crystal structure of CobB wt in complex with H4K16-Acetyl peptide | | Descriptor: | Histone H4, NAD-dependent protein deacylase, ZINC ION | | Authors: | Spinck, M, Gasper, R, Neumann, H. | | Deposit date: | 2019-06-08 | | Release date: | 2020-04-15 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Evolved, Selective Erasers of Distinct Lysine Acylations.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
