7RD8
 
 | | Structure of the S. cerevisiae P4B ATPase lipid flippase in the E1-ATP state | | Descriptor: | MAGNESIUM ION, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Probable phospholipid-transporting ATPase NEO1 | | Authors: | Bai, L, Jain, B.K, You, Q, Duan, H.D, Graham, T.R, Li, H. | | Deposit date: | 2021-07-09 | | Release date: | 2021-09-29 | | Last modified: | 2021-10-27 | | Method: | ELECTRON MICROSCOPY (5.64 Å) | | Cite: | Structural basis of the P4B ATPase lipid flippase activity.
Nat Commun, 12, 2021
|
|
7RD7
 
 | | Structure of the S. cerevisiae P4B ATPase lipid flippase in the E2P-transition state | | Descriptor: | MAGNESIUM ION, Probable phospholipid-transporting ATPase NEO1, TETRAFLUOROALUMINATE ION | | Authors: | Bai, L, Jain, B.K, You, Q, Duan, H.D, Graham, T.R, Li, H. | | Deposit date: | 2021-07-09 | | Release date: | 2021-09-29 | | Last modified: | 2021-10-27 | | Method: | ELECTRON MICROSCOPY (3.08 Å) | | Cite: | Structural basis of the P4B ATPase lipid flippase activity.
Nat Commun, 12, 2021
|
|
7RD6
 
 | | Structure of the S. cerevisiae P4B ATPase lipid flippase in the E2P state | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, Probable phospholipid-transporting ATPase NEO1 | | Authors: | Bai, L, Jain, B.K, You, Q, Duan, H.D, Graham, T.R, Li, H. | | Deposit date: | 2021-07-09 | | Release date: | 2021-09-29 | | Last modified: | 2021-10-27 | | Method: | ELECTRON MICROSCOPY (3.25 Å) | | Cite: | Structural basis of the P4B ATPase lipid flippase activity.
Nat Commun, 12, 2021
|
|
7R0I
 
 | | STRUCTURAL BASIS OF ION UPTAKE IN COPPER-TRANSPORTING P1B-TYPE ATPASES | | Descriptor: | MAGNESIUM ION, POTASSIUM ION, Putative copper-exporting P-type ATPase A | | Authors: | Salustros, N, Groenberg, C, Wang, K, Gourdon, P. | | Deposit date: | 2022-02-02 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis of ion uptake in copper-transporting P 1B -type ATPases.
Nat Commun, 13, 2022
|
|
7R0H
 
 | | STRUCTURAL BASIS OF ION UPTAKE IN COPPER-TRANSPORTING P1B-TYPE ATPASES | | Descriptor: | COPPER (II) ION, Putative copper-exporting P-type ATPase A | | Authors: | Salustros, N, Groenberg, C, Wang, K, Gourdon, P. | | Deposit date: | 2022-02-02 | | Release date: | 2022-09-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.31 Å) | | Cite: | Structural basis of ion uptake in copper-transporting P 1B -type ATPases.
Nat Commun, 13, 2022
|
|
7R0G
 
 | |
7QTV
 
 | | Beryllium fluoride form of the Na+,K+-ATPase (E2-BeFx) | | Descriptor: | 1-O-decanoyl-beta-D-tagatofuranosyl beta-D-allopyranoside, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Fruergaard, M.U, Dach, I, Andersen, J.L, Ozol, M, Shahsavar, A, Quistgaard, E.M, Poulsen, H, Fedosova, N.U, Nissen, P. | | Deposit date: | 2022-01-16 | | Release date: | 2022-11-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (4.05 Å) | | Cite: | The Na + ,K + -ATPase in complex with beryllium fluoride mimics an ATPase phosphorylated state.
J.Biol.Chem., 298, 2022
|
|
7QC0
 
 | | Crystal structure of Cadmium translocating P-type ATPase | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, Cadmium translocating P-type ATPase, MAGNESIUM ION | | Authors: | Groenberg, C, Hu, Q, Wang, K, Gourdon, P. | | Deposit date: | 2021-11-21 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.11 Å) | | Cite: | Structure and ion-release mechanism of P IB-4 -type ATPases.
Elife, 10, 2021
|
|
7QBZ
 
 | | Crystal structure Cadmium translocating P-type ATPase | | Descriptor: | Cadmium translocating P-type ATPase, MAGNESIUM ION, TETRAFLUOROALUMINATE ION | | Authors: | Groenberg, C, Hu, Q, Wang, K, Gourdon, P. | | Deposit date: | 2021-11-21 | | Release date: | 2022-03-23 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Structure and ion-release mechanism of P IB-4 -type ATPases.
Elife, 10, 2021
|
|
7PY4
 
 | | Cryo-EM structure of ATP8B1-CDC50A in E2P autoinhibited state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BERYLLIUM TRIFLUORIDE ION, CHOLESTEROL HEMISUCCINATE, ... | | Authors: | Dieudonne, T, Abad-Herrera, S, Juknaviciute Laursen, M, Lejeune, M, Stock, C, Slimani, K, Jaxel, C, Lyons, J.A, Montigny, C, Gunther Pomorski, T, Nissen, P, Lenoir, G. | | Deposit date: | 2021-10-08 | | Release date: | 2022-04-27 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Autoinhibition and regulation by phosphoinositides of ATP8B1, a human lipid flippase associated with intrahepatic cholestatic disorders.
Elife, 11, 2022
|
|
7OP8
 
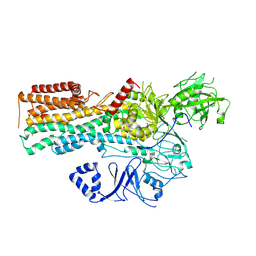 | | Cryo-EM structure of P5B-ATPase E2Pinhibit | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, Cation-transporting ATPase, MAGNESIUM ION | | Authors: | Li, P, Gourdon, P. | | Deposit date: | 2021-05-31 | | Release date: | 2021-06-30 | | Last modified: | 2021-09-01 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure and transport mechanism of P5B-ATPases.
Nat Commun, 12, 2021
|
|
7OP5
 
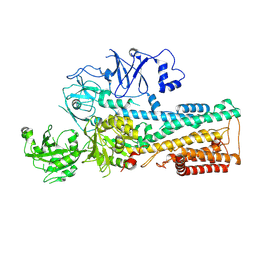 | | Cryo-EM structure of P5B-ATPase E2P | | Descriptor: | BERYLLIUM TRIFLUORIDE ION, MAGNESIUM ION, P5B-ATPase | | Authors: | Li, P, Gourdon, P. | | Deposit date: | 2021-05-29 | | Release date: | 2021-06-30 | | Last modified: | 2021-09-01 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure and transport mechanism of P5B-ATPases.
Nat Commun, 12, 2021
|
|
7OP3
 
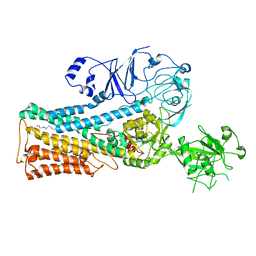 | | Cryo-EM structure of P5B-ATPase E2PiSPM | | Descriptor: | Cation-transporting ATPase, SPERMINE | | Authors: | Li, P, Gronberg, C, Wang, K.T, Salustros, N, Gourdon, P.E. | | Deposit date: | 2021-05-28 | | Release date: | 2021-06-30 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Structure and transport mechanism of P5B-ATPases.
Nat Commun, 12, 2021
|
|
7OP1
 
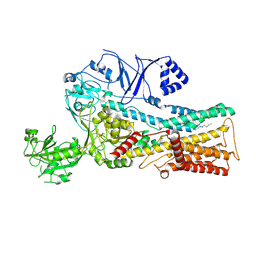 | | Cryo-EM structure of P5B-ATPase E2PiAlF/SPM | | Descriptor: | Cation-transporting ATPase, MAGNESIUM ION, SPERMINE, ... | | Authors: | Li, P, Gourdon, P. | | Deposit date: | 2021-05-28 | | Release date: | 2021-06-30 | | Last modified: | 2021-09-01 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Structure and transport mechanism of P5B-ATPases.
Nat Commun, 12, 2021
|
|
7NY1
 
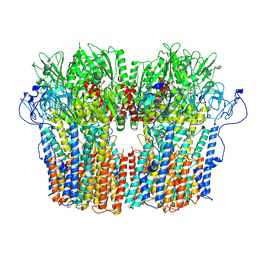 | | Structure of the fungal plasma membrane proton pump Pma1 in its auto-inhibited state - hexameric assembly | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Heit, S, Geurts, M.M.G, Murphy, B.J, Corey, R, Mills, D.J, Kuehlbrandt, W, Bublitz, M. | | Deposit date: | 2021-03-19 | | Release date: | 2021-11-17 | | Last modified: | 2021-12-01 | | Method: | ELECTRON MICROSCOPY (3.26 Å) | | Cite: | Structure of the hexameric fungal plasma membrane proton pump in its autoinhibited state.
Sci Adv, 7, 2021
|
|
7NXF
 
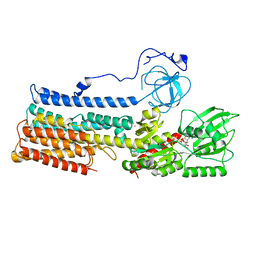 | | Structure of the fungal plasma membrane proton pump Pma1 in its auto-inhibited state - monomer unit | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Heit, S, Geurts, M.M.G, Murphy, B.J, Corey, R, Mills, D.J, Kuehlbrandt, W, Bublitz, M. | | Deposit date: | 2021-03-18 | | Release date: | 2021-11-17 | | Last modified: | 2021-12-01 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structure of the hexameric fungal plasma membrane proton pump in its autoinhibited state.
Sci Adv, 7, 2021
|
|
7NNP
 
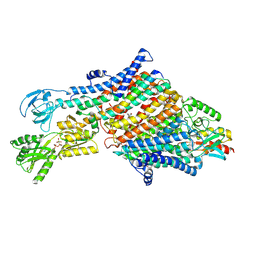 | | Rb-loaded cryo-EM structure of the E1-ATP KdpFABC complex. | | Descriptor: | CARDIOLIPIN, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, Potassium-transporting ATPase ATP-binding subunit, ... | | Authors: | Silberberg, J.M, Corey, R.A, Hielkema, L, Stock, C, Stansfeld, P.J, Paulino, C, Haenelt, I. | | Deposit date: | 2021-02-25 | | Release date: | 2021-07-28 | | Last modified: | 2021-09-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Deciphering ion transport and ATPase coupling in the intersubunit tunnel of KdpFABC.
Nat Commun, 12, 2021
|
|
7NNL
 
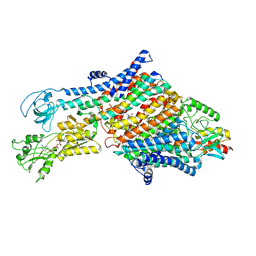 | | Cryo-EM structure of the KdpFABC complex in an E1-ATP conformation loaded with K+ | | Descriptor: | CARDIOLIPIN, PHOSPHOMETHYLPHOSPHONIC ACID ADENYLATE ESTER, POTASSIUM ION, ... | | Authors: | Silberberg, J.M, Corey, R.A, Hielkema, L, Stock, C, Stansfeld, P.J, Paulino, C, Haenelt, I. | | Deposit date: | 2021-02-25 | | Release date: | 2021-07-28 | | Last modified: | 2021-09-29 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Deciphering ion transport and ATPase coupling in the intersubunit tunnel of KdpFABC.
Nat Commun, 12, 2021
|
|
7N78
 
 | | Cryo-EM structure of ATP13A2 in the E2-Pi state | | Descriptor: | 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, CHOLESTEROL HEMISUCCINATE, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Sim, S.I, Park, E. | | Deposit date: | 2021-06-09 | | Release date: | 2021-11-10 | | Last modified: | 2021-12-01 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Structural basis of polyamine transport by human ATP13A2 (PARK9).
Mol.Cell, 81, 2021
|
|
7N77
 
 | |
7N76
 
 | |
7N75
 
 | |
7N74
 
 | |
7N73
 
 | |
7N72
 
 | |
