9C6X
 
 | | Crystal Structure of a single chain trimer composed of HLA-B*39:01 Y84C variant, beta-2microglobulin, and NRVMLPKAA peptide from NLRP2 | | Descriptor: | 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, NACHT, ... | | Authors: | Sharma, R, Amdare, N.P, Celikgil, A, Garforth, S.J, DiLorenzo, T.P, Almo, S.C, Ghosh, A. | | Deposit date: | 2024-06-09 | | Release date: | 2024-09-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and biochemical analysis of highly similar HLA-B allotypes differentially associated with type 1 diabetes.
J.Biol.Chem., 300, 2024
|
|
9C6W
 
 | | Crystal Structure of a single chain trimer composed of HLA-B*39:06 Y84C variant, beta-2microglobulin, and NRVMLPKAA peptide from NLRP2 (2 molecules/asymmetric unit) | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15-PENTAOXAHEPTADECANE, CHLORIDE ION, ... | | Authors: | Sharma, R, Amdare, N.P, Celikgil, A, Garforth, S.J, DiLorenzo, T.P, Almo, S.C, Ghosh, A. | | Deposit date: | 2024-06-09 | | Release date: | 2024-09-04 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural and biochemical analysis of highly similar HLA-B allotypes differentially associated with type 1 diabetes.
J.Biol.Chem., 300, 2024
|
|
9DH3
 
 | | Cryo-EM structure of NLRP3 complex with Compound C | | Descriptor: | 2-[(4S)-5-ethyl-8-oxothieno[2',3':4,5]pyrrolo[1,2-d][1,2,4]triazin-7(8H)-yl]-N-(pyrimidin-4-yl)acetamide, 3-[(3-CHOLAMIDOPROPYL)DIMETHYLAMMONIO]-1-PROPANESULFONATE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Matico, R, Grauwen, K, Van Gool, M, Muratore, E.M, Yu, X, Abdiaj, I, Adhikary, S, Adriaensen, I, Aranzazu, G.M, Alcazar, J, Bassi, M, Brisse, E, Canellas, S, Chaudhuri, S, Chauhan, D, Delgado, F, Dieguez-Vazquez, A, Du Jardin, M, Eastham, V, Finley, M, Jacobs, T, Keustermans, K, Kuhn, R, Llaveria, J, Leenaerts, J, Linares, M.L, Martin, M.L, Martinez, C, Miller, R, Munoz, F.M, Nooyens, A, Perez, L.B, Perrier, M, Pietrak, B, Serre, J, Sharma, S, Somers, M, Suarez, J, Tresadern, G, Trabanco, A.A, Van den Bulck, D, Van Hauwermeiren, F, Varghese, T, Vega, J.A, Youssef, S.A, Edwards, M.J, Oehlrich, D, Van Opdenbosch, N. | | Deposit date: | 2024-09-03 | | Release date: | 2024-12-25 | | Last modified: | 2025-01-22 | | Method: | ELECTRON MICROSCOPY (3.76 Å) | | Cite: | Navigating from cellular phenotypic screen to clinical candidate: selective targeting of the NLRP3 inflammasome.
Embo Mol Med, 17, 2025
|
|
5JUY
 
 | | Active human apoptosome with procaspase-9 | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Apoptotic protease-activating factor 1, Caspase-9, ... | | Authors: | Cheng, T.C, Hong, C, Akey, I.V, Yuan, S, Akey, C.W. | | Deposit date: | 2016-05-10 | | Release date: | 2016-10-19 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | A near atomic structure of the active human apoptosome.
Elife, 5, 2016
|
|
4RHW
 
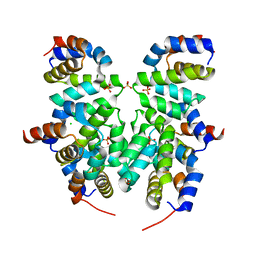 | | Crystal structure of Apaf-1 CARD and caspase-9 CARD complex | | Descriptor: | Apoptotic protease-activating factor 1, CHLORIDE ION, Caspase-9, ... | | Authors: | Hu, Q, Wu, D, Yan, C, Shi, Y. | | Deposit date: | 2014-10-03 | | Release date: | 2014-10-15 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Molecular determinants of caspase-9 activation by the Apaf-1 apoptosome.
Proc. Natl. Acad. Sci. U.S.A., 111, 2014
|
|
6XFO
 
 | |
6O5J
 
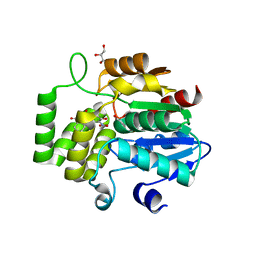 | | Crystal Structure of DAD2 bound to quinazolinone derivative | | Descriptor: | 1-(4-hydroxy-3-nitrophenyl)quinazoline-2,4(1H,3H)-dione, ACETATE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Hamiaux, C. | | Deposit date: | 2019-03-03 | | Release date: | 2019-06-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.63 Å) | | Cite: | Chemical synthesis and characterization of a new quinazolinedione competitive antagonist for strigolactone receptors with an unexpected binding mode.
Biochem.J., 476, 2019
|
|
1JXQ
 
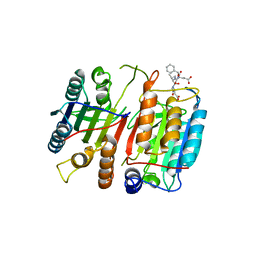 | | Structure of cleaved, CARD domain deleted Caspase-9 | | Descriptor: | Caspase-9, benzoxycarbonyl-Val-Ala-Asp-fluoromethyl ketone Inhibitor | | Authors: | Renatus, M, Stennicke, H.R, Scott, F.L, Liddington, R.C, Salvesen, G.S. | | Deposit date: | 2001-09-08 | | Release date: | 2001-12-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Dimer formation drives the activation of the cell death protease caspase 9.
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1GCC
 
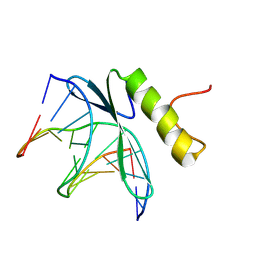 | | SOLUTION NMR STRUCTURE OF THE COMPLEX OF GCC-BOX BINDING DOMAIN OF ATERF1 AND GCC-BOX DNA, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | DNA (5'-D(*GP*CP*TP*GP*GP*CP*GP*GP*CP*TP*A)-3'), DNA (5'-D(*TP*AP*GP*CP*CP*GP*CP*CP*AP*GP*C)-3'), ETHYLENE RESPONSIVE ELEMENT BINDING FACTOR 1 | | Authors: | Yamasaki, K, Allen, M.D, Ohme-Takagi, M, Tateno, M, Suzuki, M. | | Deposit date: | 1998-03-13 | | Release date: | 1999-03-23 | | Last modified: | 2024-10-23 | | Method: | SOLUTION NMR | | Cite: | A novel mode of DNA recognition by a beta-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA.
EMBO J., 17, 1998
|
|
3SFZ
 
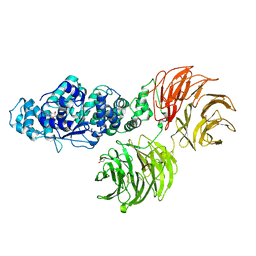 | | Crystal structure of full-length murine Apaf-1 | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Apoptotic peptidase activating factor 1, GAMMA-BUTYROLACTONE | | Authors: | Eschenburg, S, Reubold, T.F. | | Deposit date: | 2011-06-14 | | Release date: | 2011-08-24 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of full-length Apaf-1: how the death signal is relayed in the mitochondrial pathway of apoptosis.
Structure, 19, 2011
|
|
3SHF
 
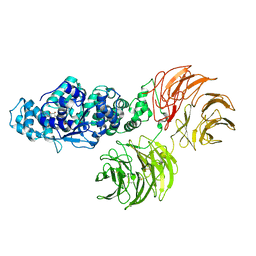 | |
6UH8
 
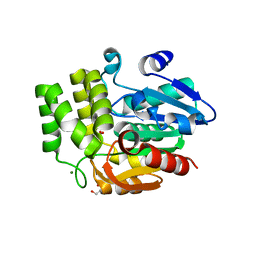 | | Crystal structure of DAD2 N242I mutant | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Decreased Apical Dominance 2, GLYCEROL, ... | | Authors: | Sharma, P, Hamiaux, C, Snowden, K.C. | | Deposit date: | 2019-09-27 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Flexibility of the petunia strigolactone receptor DAD2 promotes its interaction with signaling partners.
J.Biol.Chem., 295, 2020
|
|
2P1H
 
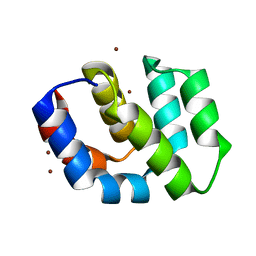 | | Rapid Folding and Unfolding of Apaf-1 CARD | | Descriptor: | Apoptotic protease-activating factor 1, ZINC ION | | Authors: | Milam, S.L, Nicely, N.I, Feeney, B, Mattos, C, Clark, A.C. | | Deposit date: | 2007-03-05 | | Release date: | 2007-05-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Rapid Folding and Unfolding of Apaf-1 CARD.
J.Mol.Biol., 369, 2007
|
|
6UH9
 
 | | Crystal structure of DAD2 D166A mutant | | Descriptor: | Decreased Apical Dominance 2, TETRAETHYLENE GLYCOL | | Authors: | Sharma, P, Hamiaux, C, Snowden, K.C. | | Deposit date: | 2019-09-27 | | Release date: | 2020-02-26 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Flexibility of the petunia strigolactone receptor DAD2 promotes its interaction with signaling partners.
J.Biol.Chem., 295, 2020
|
|
5WVE
 
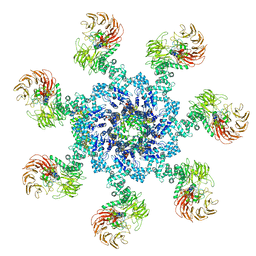 | | Apaf-1-Caspase-9 holoenzyme | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Apoptotic protease-activating factor 1, Caspase, ... | | Authors: | Li, Y, Zhou, M, Hu, Q, Shi, Y. | | Deposit date: | 2016-12-24 | | Release date: | 2017-02-08 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (4.4 Å) | | Cite: | Mechanistic insights into caspase-9 activation by the structure of the apoptosome holoenzyme
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
5WVC
 
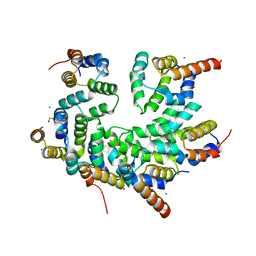 | | Structure of the CARD-CARD disk | | Descriptor: | Apoptotic protease-activating factor 1, Caspase, IODIDE ION | | Authors: | Lin, S.C, Lo, Y.C, Su, T.W. | | Deposit date: | 2016-12-24 | | Release date: | 2017-04-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.993 Å) | | Cite: | Structural Insights into DD-Fold Assembly and Caspase-9 Activation by the Apaf-1 Apoptosome.
Structure, 25, 2017
|
|
2GCC
 
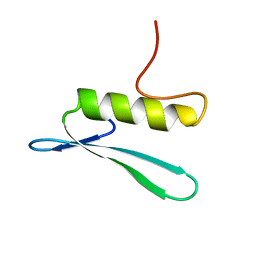 | | SOLUTION STRUCTURE OF THE GCC-BOX BINDING DOMAIN, NMR, MINIMIZED MEAN STRUCTURE | | Descriptor: | ATERF1 | | Authors: | Allen, M.D, Yamasaki, K, Ohme-Takagi, M, Tateno, M, Suzuki, M. | | Deposit date: | 1998-03-13 | | Release date: | 1999-03-23 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | A novel mode of DNA recognition by a beta-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA.
EMBO J., 17, 1998
|
|
3J2T
 
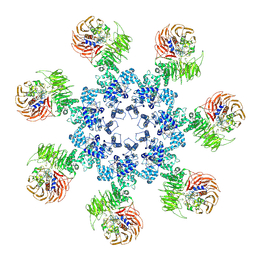 | | An improved model of the human apoptosome | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Apoptotic protease-activating factor 1, Cytochrome c, ... | | Authors: | Yuan, S, Topf, M, Akey, C.W. | | Deposit date: | 2012-12-23 | | Release date: | 2013-04-10 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (9.5 Å) | | Cite: | Changes in apaf-1 conformation that drive apoptosome assembly.
Biochemistry, 52, 2013
|
|
3GCC
 
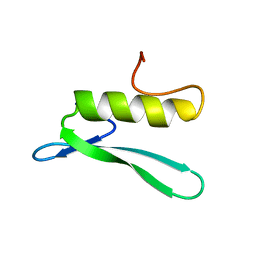 | | SOLUTION STRUCTURE OF THE GCC-BOX BINDING DOMAIN, NMR, 46 STRUCTURES | | Descriptor: | ATERF1 | | Authors: | Allen, M.D, Yamasaki, K, Ohme-Takagi, M, Tateno, M, Suzuki, M. | | Deposit date: | 1998-03-13 | | Release date: | 1999-03-23 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | A novel mode of DNA recognition by a beta-sheet revealed by the solution structure of the GCC-box binding domain in complex with DNA.
EMBO J., 17, 1998
|
|
3JBT
 
 | | Atomic structure of the Apaf-1 apoptosome | | Descriptor: | 2'-DEOXYADENOSINE 5'-TRIPHOSPHATE, Apoptotic protease-activating factor 1, Cytochrome c, ... | | Authors: | Zhou, M, Li, Y, Hu, Q, Bai, X, Huang, W, Yan, C, Scheres, S.H.W, Shi, Y. | | Deposit date: | 2015-10-15 | | Release date: | 2015-11-18 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Atomic structure of the apoptosome: mechanism of cytochrome c- and dATP-mediated activation of Apaf-1
Genes Dev., 29, 2015
|
|
2LXW
 
 | |
8XQK
 
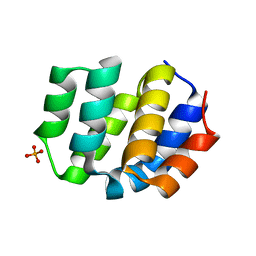 | | The Crystal Structure of Apaf from Biortus. | | Descriptor: | Apoptotic protease-activating factor 1, PHOSPHATE ION | | Authors: | Wang, F, Cheng, W, Lv, Z, Ju, C, Ni, C. | | Deposit date: | 2024-01-05 | | Release date: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | The Crystal Structure of Apaf from Biortus.
To Be Published
|
|
6AP7
 
 | |
6AP6
 
 | | Crystal Structure of DAD2 in complex with tolfenamic acid | | Descriptor: | 2-[(3-chloro-2-methylphenyl)amino]benzoic acid, Probable strigolactone esterase DAD2 | | Authors: | Hamiaux, C. | | Deposit date: | 2017-08-17 | | Release date: | 2018-03-21 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Inhibition of strigolactone receptors byN-phenylanthranilic acid derivatives: Structural and functional insights.
J. Biol. Chem., 293, 2018
|
|
6AP8
 
 | |
