6WYO
 
 | | Crystal structure of Danio rerio histone deacetylase 6 catalytic domain 1 (CD1) H82F F202Y double mutant complexed with Trichostatin A | | Descriptor: | Histone deacetylase 6, POTASSIUM ION, TRICHOSTATIN A, ... | | Authors: | Osko, J.D, Christianson, D.W. | | Deposit date: | 2020-05-13 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.30000281 Å) | | Cite: | Binding of inhibitors to active-site mutants of CD1, the enigmatic catalytic domain of histone deacetylase 6.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
6WYQ
 
 | | Crystal structure of Danio rerio histone deacetylase 6 catalytic domain 1 (CD1) K330L mutant complexed with 4-iodo-SAHA | | Descriptor: | Histone deacetylase 6, N~1~-hydroxy-N~8~-(4-iodophenyl)octanediamide, POTASSIUM ION, ... | | Authors: | Osko, J.D, Christianson, D.W. | | Deposit date: | 2020-05-13 | | Release date: | 2020-09-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.90001464 Å) | | Cite: | Binding of inhibitors to active-site mutants of CD1, the enigmatic catalytic domain of histone deacetylase 6.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
5W5K
 
 | | Crystal structure of Danio rerio histone deacetylase 6 catalytic domain 2 in complex with KV70 | | Descriptor: | 10-{[4-(hydroxycarbamoyl)phenyl]methyl}-5lambda~4~-pyrido[3,2-b][1,4]benzothiazin-10-ium, Histone deacetylase 6, POTASSIUM ION, ... | | Authors: | Porter, N.J, Christianson, D.W. | | Deposit date: | 2017-06-15 | | Release date: | 2018-06-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Synthesis and Biological Investigation of Phenothiazine-Based Benzhydroxamic Acids as Selective Histone Deacetylase 6 Inhibitors.
J.Med.Chem., 62, 2019
|
|
2VCG
 
 | | Crystal structure of a HDAC-like protein HDAH from Bordetella sp. with the bound inhibitor ST-17 | | Descriptor: | CHLORIDE ION, GLYCEROL, HISTONE DEACETYLASE-LIKE AMIDOHYDROLASE, ... | | Authors: | Dickmanns, A, Strasser, A, Ficner, R. | | Deposit date: | 2007-09-24 | | Release date: | 2008-01-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Phenylalanine-Containing Hydroxamic Acids as Selective Inhibitors of Class Iib Histone Deacetylases (Hdacs).
Bioorg.Med.Chem., 16, 2008
|
|
7U69
 
 | |
7U3M
 
 | |
7U6B
 
 | |
7U6A
 
 | |
6WHO
 
 | | Histone deacetylases complex with peptide macrocycles | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Histone deacetylase 2, SODIUM ION, ... | | Authors: | Bera, A.K, Hosseinzadeh, P, Watson, P, Baker, D. | | Deposit date: | 2020-04-08 | | Release date: | 2021-04-21 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Anchor extension: a structure-guided approach to design cyclic peptides targeting enzyme active sites.
Nat Commun, 12, 2021
|
|
6WHN
 
 | | Histone deacetylases complex with peptide macrocycles | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, DI(HYDROXYETHYL)ETHER, Histone deacetylase 2, ... | | Authors: | Bera, A.K, Hosseinzadeh, P, Watson, P, Baker, D. | | Deposit date: | 2020-04-08 | | Release date: | 2021-04-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Anchor extension: a structure-guided approach to design cyclic peptides targeting enzyme active sites.
Nat Commun, 12, 2021
|
|
6WHQ
 
 | | Histone deacetylases complex with peptide macrocycles | | Descriptor: | 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Histone deacetylase 2, SODIUM ION, ... | | Authors: | Bera, A.K, Hosseinzadeh, P, Watson, P, Baker, D. | | Deposit date: | 2020-04-08 | | Release date: | 2021-04-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Anchor extension: a structure-guided approach to design cyclic peptides targeting enzyme active sites.
Nat Commun, 12, 2021
|
|
6WHZ
 
 | | Histone deacetylases complex with peptide macrocycles | | Descriptor: | Histone deacetylase 2, SODIUM ION, TETRAETHYLENE GLYCOL, ... | | Authors: | Bera, A.K, Hosseinzadeh, P, Watson, P, Baker, D. | | Deposit date: | 2020-04-08 | | Release date: | 2021-04-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Anchor extension: a structure-guided approach to design cyclic peptides targeting enzyme active sites.
Nat Commun, 12, 2021
|
|
6WI3
 
 | | Histone deacetylases complex with peptide macrocycles | | Descriptor: | (SHA)W(DTH)DN(DSN)(DME)(DAS)K peptide macrocycle, 2-[N-CYCLOHEXYLAMINO]ETHANE SULFONIC ACID, Histone deacetylase 2, ... | | Authors: | Bera, A.K, Hosseinzadeh, P, Watson, P, Baker, D. | | Deposit date: | 2020-04-08 | | Release date: | 2021-04-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Anchor extension: a structure-guided approach to design cyclic peptides targeting enzyme active sites.
Nat Commun, 12, 2021
|
|
5VI6
 
 | | Crystal structure of histone deacetylase 8 in complex with trapoxin A | | Descriptor: | 1,2-ETHANEDIOL, 1,4-DIETHYLENE DIOXIDE, Histone deacetylase 8, ... | | Authors: | Porter, N.J, Christianson, D.W. | | Deposit date: | 2017-04-14 | | Release date: | 2017-09-06 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.237 Å) | | Cite: | Binding of the Microbial Cyclic Tetrapeptide Trapoxin A to the Class I Histone Deacetylase HDAC8.
ACS Chem. Biol., 12, 2017
|
|
6H39
 
 | |
2I50
 
 | | Solution Structure of Ubp-M Znf-UBP domain | | Descriptor: | Ubiquitin carboxyl-terminal hydrolase 16, ZINC ION | | Authors: | Pai, M.-T. | | Deposit date: | 2006-08-23 | | Release date: | 2007-08-07 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Ubp-M BUZ domain, a highly specific protein module that recognizes the C-terminal tail of free ubiquitin.
J.Mol.Biol., 370, 2007
|
|
7O2V
 
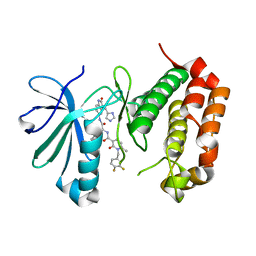 | | AURORA KINASE A IN COMPLEX WITH THE AUR-A/PDK1 INHIBITOR VI8 | | Descriptor: | 1-[[3,4-bis(fluoranyl)phenyl]methyl]-~{N}-[(1~{R})-2-[[(3~{E})-3-(1~{H}-imidazol-5-ylmethylidene)-2-oxidanylidene-1~{H}-indol-5-yl]amino]-2-oxidanylidene-1-phenyl-ethyl]-6-methyl-2-oxidanylidene-pyridine-3-carboxamide, Aurora kinase A | | Authors: | Garau, G. | | Deposit date: | 2021-03-31 | | Release date: | 2021-10-20 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Development of potent dual PDK1/AurA kinase inhibitors for cancer therapy: Lead-optimization, structural insights, and ADME-Tox profile.
Eur.J.Med.Chem., 226, 2021
|
|
7KUS
 
 | | Crystal Structure of Danio rerio Histone Deacetylase 10 H137A Mutant in Complex with N8-Acetylspermidine (Tetrahedral Intermediate) | | Descriptor: | 1,2-ETHANEDIOL, 1-({4-[(3-aminopropyl)amino]butyl}amino)ethane-1,1-diol, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Herbst-Gervasoni, C.J, Christianson, D.W. | | Deposit date: | 2020-11-25 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-ray Crystallographic Snapshots of Substrate Binding in the Active Site of Histone Deacetylase 10.
Biochemistry, 60, 2021
|
|
7KUQ
 
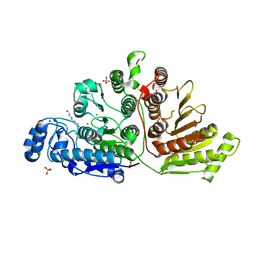 | |
7KUR
 
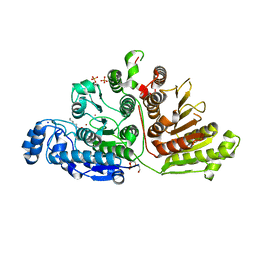 | |
7KUT
 
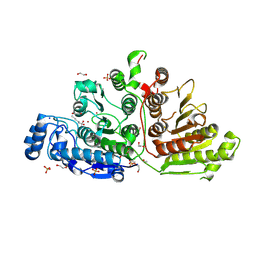 | | Crystal Structure of Danio rerio Histone Deacetylase 10 H137A Mutant in Complex with N-Acetylputrescine (Tetrahedral Intermediate) | | Descriptor: | 1,2-ETHANEDIOL, 1-[(4-aminobutyl)amino]ethane-1,1-diol, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Herbst-Gervasoni, C.J, Christianson, D.W. | | Deposit date: | 2020-11-25 | | Release date: | 2021-02-03 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | X-ray Crystallographic Snapshots of Substrate Binding in the Active Site of Histone Deacetylase 10.
Biochemistry, 60, 2021
|
|
7KUV
 
 | |
6FYZ
 
 | | Development and characterization of a CNS-penetrant benzhydryl hydroxamic acid class IIa histone deacetylase inhibitor | | Descriptor: | (2~{S})-2-(2-fluorophenyl)-2-[4-(2-methylpyrimidin-5-yl)phenyl]-~{N}-oxidanyl-ethanamide, Histone deacetylase 4, SODIUM ION, ... | | Authors: | Luckhurst, C.A, Maillard, M.C, Dominguez, C. | | Deposit date: | 2018-03-13 | | Release date: | 2018-12-05 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Development and characterization of a CNS-penetrant benzhydryl hydroxamic acid class IIa histone deacetylase inhibitor.
Bioorg. Med. Chem. Lett., 29, 2019
|
|
8OUA
 
 | | Cereblon isoform 4 in complex with novel Benzamide-Type Cereblon Binder 11f | | Descriptor: | 4-azanyl-~{N}-[(3~{S})-2,6-bis(oxidanylidene)piperidin-3-yl]-2,5-bis(fluoranyl)-3-methoxy-benzamide, Cereblon isoform 4, PHOSPHATE ION, ... | | Authors: | Heim, C, Bischof, L, Hartmann, M.D. | | Deposit date: | 2023-04-22 | | Release date: | 2023-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Leveraging Ligand Affinity and Properties: Discovery of Novel Benzamide-Type Cereblon Binders for the Design of PROTACs.
J.Med.Chem., 66, 2023
|
|
8OU4
 
 | | Cereblon isoform 4 in complex with novel Benzamide-Type Cereblon Binder 11a | | Descriptor: | 4-azanyl-~{N}-[(3~{S})-2,6-bis(oxidanylidene)piperidin-3-yl]-2-chloranyl-benzamide, Cereblon isoform 4, ZINC ION | | Authors: | Heim, C, Bischof, L, Hartmann, M.D. | | Deposit date: | 2023-04-21 | | Release date: | 2023-11-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Leveraging Ligand Affinity and Properties: Discovery of Novel Benzamide-Type Cereblon Binders for the Design of PROTACs.
J.Med.Chem., 66, 2023
|
|
