1GRB
 
 | |
8JWO
 
 | |
1H2A
 
 | | SINGLE CRYSTALS OF HYDROGENASE FROM DESULFOVIBRIO VULGARIS | | Descriptor: | FE3-S4 CLUSTER, HYDROGENASE, IRON/SULFUR CLUSTER, ... | | Authors: | Higuchi, Y, Yasuoka, N. | | Deposit date: | 1997-10-17 | | Release date: | 1999-02-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Unusual ligand structure in Ni-Fe active center and an additional Mg site in hydrogenase revealed by high resolution X-ray structure analysis.
Structure, 5, 1997
|
|
8JWL
 
 | |
8JWK
 
 | | The second purified state crystal structure of AKRtyl | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, Aldo/keto reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Lin, S, Dai, S, Xiao, Z. | | Deposit date: | 2023-06-29 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | A three-level regulatory mechanism of the aldo-keto reductase subfamily AKR12D.
Nat Commun, 15, 2024
|
|
8JWM
 
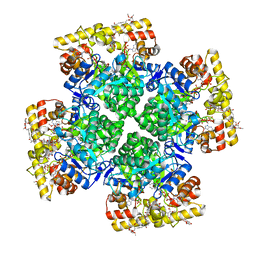 | | Crystal structure of AKRtyl-NADP-tylosin complex | | Descriptor: | Aldo/keto reductase, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, TYLOSIN | | Authors: | Lin, S, Dai, S, Xiao, Z. | | Deposit date: | 2023-06-29 | | Release date: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | A three-level regulatory mechanism of the aldo-keto reductase subfamily AKR12D.
Nat Commun, 15, 2024
|
|
2LLS
 
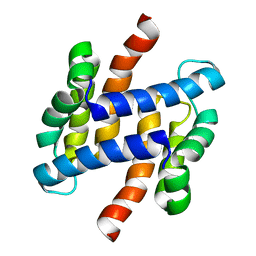 | | solution structure of human apo-S100A1 C85M | | Descriptor: | Protein S100-A1 | | Authors: | Budzinska, M, Jaremko, L, Jaremko, M, Zdanowski, K, Zhukov, I, Bierzynski, A, Ejchart, A. | | Deposit date: | 2011-11-17 | | Release date: | 2012-12-19 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Chemical Shift Assignments and solution structure of human apo-S100A1 C85M mutant
To be Published
|
|
2LQW
 
 | |
1I7K
 
 | |
1HRA
 
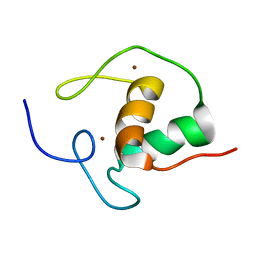 | | THE SOLUTION STRUCTURE OF THE HUMAN RETINOIC ACID RECEPTOR-BETA DNA-BINDING DOMAIN | | Descriptor: | RETINOIC ACID RECEPTOR, ZINC ION | | Authors: | Knegtel, R.M.A, Katahira, M, Schilthuis, J.G, Bonvin, A.M.J.J, Boelens, R, Eib, D, Van Der Saag, P.T, Kaptein, R. | | Deposit date: | 1993-07-25 | | Release date: | 1994-01-31 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | The solution structure of the human retinoic acid receptor-beta DNA-binding domain.
J.Biomol.NMR, 3, 1993
|
|
2L7J
 
 | |
8SOR
 
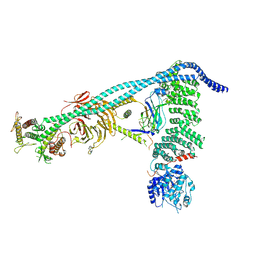 | | Structure of human PI3KC3-C1 complex | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Beclin 1-associated autophagy-related key regulator, Beclin-1, ... | | Authors: | Chen, M, Hurley, J.H. | | Deposit date: | 2023-04-29 | | Release date: | 2023-06-21 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.96 Å) | | Cite: | Structure and activation of the human autophagy-initiating ULK1C:PI3KC3-C1 supercomplex
bioRxiv, 2023
|
|
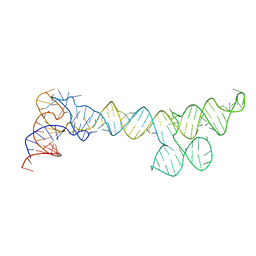
8SP9
 
 | |
2LUC
 
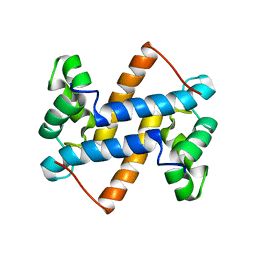 | |
8SYL
 
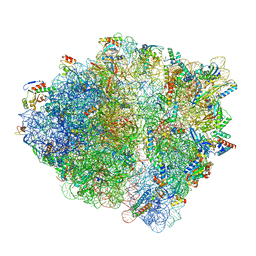 | | Cryo-EM structure of the Escherichia coli 70S ribosome in complex with amikacin, mRNA, and A-, P-, and E-site tRNAs | | Descriptor: | (2S)-N-[(1R,2S,3S,4R,5S)-4-[(2R,3R,4S,5S,6R)-6-(aminomethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]oxy-5-azanyl-2-[(2S,3R,4S,5S ,6R)-4-azanyl-6-(hydroxymethyl)-3,5-bis(oxidanyl)oxan-2-yl]oxy-3-oxidanyl-cyclohexyl]-4-azanyl-2-oxidanyl-butanamide, 16S Ribosomal RNA, 23S Ribosomal RNA, ... | | Authors: | Seely, S.M, Gagnon, M.G. | | Deposit date: | 2023-05-25 | | Release date: | 2023-08-09 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular basis of the pleiotropic effects by the antibiotic amikacin on the ribosome.
Nat Commun, 14, 2023
|
|
8KEM
 
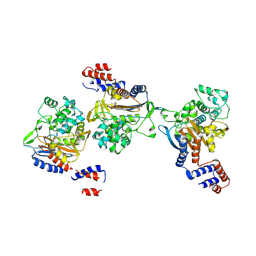 | | PKS domains-fused AmpC EC2 | | Descriptor: | SDR family NAD(P)-dependent oxidoreductase,Beta-lactamase | | Authors: | Son, S.Y, Bae, D.W, Cha, S.S. | | Deposit date: | 2023-08-12 | | Release date: | 2024-06-05 | | Last modified: | 2024-09-18 | | Method: | X-RAY DIFFRACTION (2.76998281 Å) | | Cite: | Structural investigation of the docking domain assembly from trans-AT polyketide synthases.
Structure, 32, 2024
|
|
2MFQ
 
 | |
1HTR
 
 | |
2JKK
 
 | |
8JV3
 
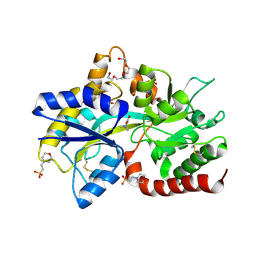 | |
8JV4
 
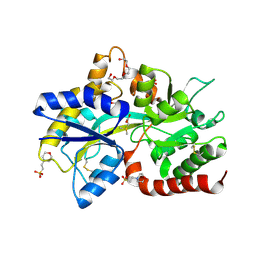 | | Structure of the SAR11 PotD in complex with DMSP | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-(dimethyl-lambda~4~-sulfanyl)propanoic acid, GLYCEROL, ... | | Authors: | Ma, Q, Liu, C. | | Deposit date: | 2023-06-27 | | Release date: | 2024-07-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.445 Å) | | Cite: | Structure of the SAR11 PotD in complex with DMSP
To Be Published
|
|
8JV1
 
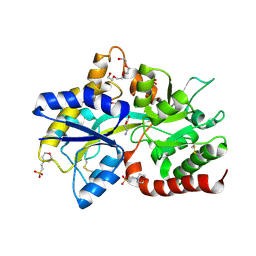 | | Structure of the SAR11 PotD in complex with GABA | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CHLORIDE ION, GAMMA-AMINO-BUTANOIC ACID, ... | | Authors: | Ma, Q, Liu, C. | | Deposit date: | 2023-06-27 | | Release date: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (1.309 Å) | | Cite: | Structure of the SAR11 PotD in complex with GABA
To Be Published
|
|
7ZWB
 
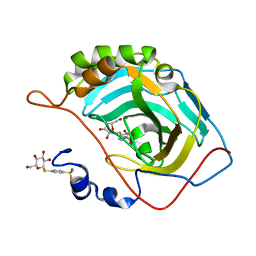 | | human Carbonic Anhydrase II in complex with 4-(((2S,3R,4S,5S,6R)-3,4,5-trihydroxy-6-(hydroxymethyl)tetrahydro-2H-pyran-2-yl)thio)benzenesulfonamide | | Descriptor: | 4-[(2~{S},3~{R},4~{S},5~{S},6~{R})-6-(hydroxymethyl)-3,4,5-tris(oxidanyl)oxan-2-yl]sulfanylbenzenesulfonamide, Carbonic anhydrase 2, ZINC ION | | Authors: | Angeli, A, Ferraroni, M. | | Deposit date: | 2022-05-19 | | Release date: | 2023-05-31 | | Last modified: | 2024-06-12 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | First-in-Class Dual Targeting Compounds for the Management of Seizures in Glucose Transporter Type 1 Deficiency Syndrome.
J.Med.Chem., 66, 2023
|
|
8JV2
 
 | | Structure of the SAR11 PotD in complex with proline | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, GLYCEROL, PROLINE, ... | | Authors: | Ma, Q, Liu, C. | | Deposit date: | 2023-06-27 | | Release date: | 2024-07-03 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Structure of the SAR11 PotD in complex with proline
To Be Published
|
|
2EZP
 
 | |
