4W2R
 
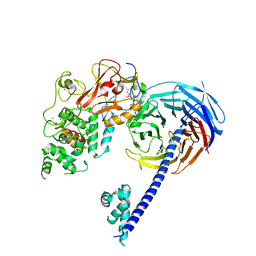 | | Structure of Hs/AcPRC2 in complex with 5,8-dichloro-2-[(4-methoxy-6-methyl-2-oxo-1,2-dihydropyridin-3-yl)methyl]-7-[(R)-methoxy(oxetan-3-yl)methyl]-3,4-dihydroisoquinolin-1(2H)-one | | Descriptor: | 5,8-dichloro-2-[(4-methoxy-6-methyl-2-oxo-1,2-dihydropyridin-3-yl)methyl]-7-[(R)-methoxy(oxetan-3-yl)methyl]-3,4-dihydroisoquinolin-1(2H)-one, Enhancer of zeste 2 polycomb repressive complex 2 subunit, Polycomb protein EED, ... | | Authors: | Gajiwala, K.S, Brooun, A, Liu, W, Deng, Y, Stewart, A.E. | | Deposit date: | 2017-09-25 | | Release date: | 2017-12-27 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Optimization of Orally Bioavailable Enhancer of Zeste Homolog 2 (EZH2) Inhibitors Using Ligand and Property-Based Design Strategies: Identification of Development Candidate (R)-5,8-Dichloro-7-(methoxy(oxetan-3-yl)methyl)-2-((4-methoxy-6-methyl-2-oxo-1,2-dihydropyridin-3-yl)methyl)-3,4-dihydroisoquinolin-1(2H)-one (PF-06821497).
J. Med. Chem., 61, 2018
|
|
4V0L
 
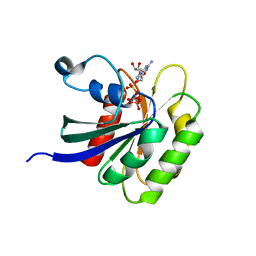 | |
2UVO
 
 | | High Resolution Crystal Structure of Wheat Germ Agglutinin in Complex with N-Acetyl-D-Glucosamine | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, AGGLUTININ ISOLECTIN 1, ... | | Authors: | Schwefel, D, Wittmann, V, Diederichs, K, Welte, W. | | Deposit date: | 2007-03-13 | | Release date: | 2008-05-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural Basis of Multivalent Binding to Wheat Germ Agglutinin.
J.Am.Chem.Soc., 132, 2010
|
|
8H6A
 
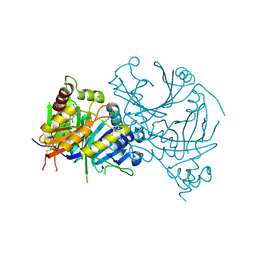 | | Crystal structure of AtHPPD complexed with YH21477 | | Descriptor: | 1-[(2-chlorophenyl)methyl]-3-methyl-N-(1-methyl-1,2,3,4-tetrazol-5-yl)-2-oxidanylidene-benzimidazole-4-carboxamide, 4-hydroxyphenylpyruvate dioxygenase, COBALT (II) ION | | Authors: | Yang, G.-F. | | Deposit date: | 2022-10-16 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.601 Å) | | Cite: | Crystal structure of AtHPPD complexed with YH20702
To Be Published
|
|
8HOD
 
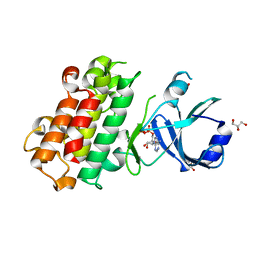 | | ScRIPK MUTANT-S253A, T254A | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GLYCEROL, Hypothetical protein | | Authors: | Fang, J.L, Zhang, M.Q. | | Deposit date: | 2022-12-09 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Receptor-like cytoplasmic kinase ScRIPK in sugarcane regulates disease resistance and drought tolerance in Arabidopsis.
Front Plant Sci, 14, 2023
|
|
2UU8
 
 | | X-ray structure of Ni, Ca concanavalin A at Ultra-high resolution (0. 94A) | | Descriptor: | CALCIUM ION, CONCANAVALIN, NICKEL (II) ION | | Authors: | Ahmed, H.U, Blakeley, M.P, Cianci, M, Cruickshank, D.W.J, Hubbard, J.A, Helliwell, J.R. | | Deposit date: | 2007-03-01 | | Release date: | 2007-07-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (0.94 Å) | | Cite: | The Determination of Protonation States in Proteins.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
4UTU
 
 | | Structural and biochemical characterization of the N- acetylmannosamine-6-phosphate 2-epimerase from Clostridium perfringens | | Descriptor: | CHLORIDE ION, N-ACETYLMANNOSAMINE-6-PHOSPHATE 2-EPIMERASE, N-acetylmannosamine-6-phosphate | | Authors: | Pelissier, M.C, Sebban-Kreuzer, C, Guerlesquin, F, Brannigan, J.A, Davies, G.J, Bourne, Y, Vincent, F. | | Deposit date: | 2014-07-23 | | Release date: | 2014-10-15 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural and Functional Characterization of the Clostridium Perfringens N-Acetylmannosamine-6-Phosphate 2-Epimerase Essential for the Sialic Acid Salvage Pathway
J.Biol.Chem., 289, 2014
|
|
4UWM
 
 | | Type II Baeyer-Villiger monooxygenase.The oxygenating constituent of 3,6-diketocamphane monooxygenase from CAM plasmid of Pseudomonas putida in complex with FMN. | | Descriptor: | 3,6-DIKETOCAMPHANE 1,6 MONOOXYGENASE, DI(HYDROXYETHYL)ETHER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Isupov, M.N, Schroeder, E, Gibson, R.P, Beecher, J, Donadio, G, Saneei, V, Dcunha, S, McGhie, E.J, Sayer, C, Davenport, C.F, Lau, P.C, Hasegawa, Y, Iwaki, H, Kadow, M, Loschinski, K, Bornscheuer, U.T, Bourenkov, G, Littlechild, J.A. | | Deposit date: | 2014-08-12 | | Release date: | 2015-08-26 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The Oxygenating Constituent of 3,6-Diketocamphane Monooxygenase from the Cam Plasmid of Pseudomonas Putida: The First Crystal Structure of a Type II Baeyer-Villiger Monooxygenase.
Acta Crystallogr.,Sect.D, 71, 2015
|
|
2UU9
 
 | | Structure of the Thermus thermophilus 30S ribosomal subunit complexed with a Valine-ASL with cmo5U in position 34 bound to an mRNA with a GUG-codon in the A-site and paromomycin. | | Descriptor: | 16S RRNA, 30S RIBOSOMAL PROTEIN S10, 30S RIBOSOMAL PROTEIN S11, ... | | Authors: | Weixlbaumer, A, Murphy, F.V, Dziergowska, A, Malkiewicz, A, Vendeix, F.A.P, Agris, P.F, Ramakrishnan, V. | | Deposit date: | 2007-03-01 | | Release date: | 2007-05-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Mechanism for expanding the decoding capacity of transfer RNAs by modification of uridines.
Nat. Struct. Mol. Biol., 14, 2007
|
|
8D9S
 
 | | AP-1, Arf1, Nef lattice on MHC-I lipopeptide incorporated wide membrane tubes, centered on beta-Arf1 | | Descriptor: | ADP ribosylation factor 1, AP-1 complex subunit beta-1, AP-1 complex subunit gamma-1, ... | | Authors: | Hooy, R.M, Hurley, J.H. | | Deposit date: | 2022-06-11 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Self-assembly and structure of a clathrin-independent AP-1:Arf1 tubular membrane coat.
Sci Adv, 8, 2022
|
|
4UZD
 
 | | SAR156497 an exquisitely selective inhibitor of Aurora kinases | | Descriptor: | AURORA KINASE A, ethyl (9S)-9-[3-(1H-benzimidazol-2-yloxy)phenyl]-8-oxo-4,5,6,7,8,9-hexahydro-2H-pyrrolo[3,4-b]quinoline-3-carboxylate | | Authors: | Pouzieux, S, Delarbre, L, Crenne, J.Y. | | Deposit date: | 2014-09-05 | | Release date: | 2014-11-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Sar156497 an Exquisitely Selective Inhibitor of Aurora Kinases.
J.Med.Chem., 58, 2015
|
|
2UW4
 
 | | Structure of PKA-PKB chimera complexed with 2-(4-(5-methyl-1H-pyrazol- 4-yl)-phenyl)-ethylamine | | Descriptor: | 2-[4-(3-METHYL-1H-PYRAZOL-4-YL)PHENYL]ETHANAMINE, CAMP-DEPENDENT PROTEIN KINASE INHIBITOR ALPHA, CAMP-DEPENDENT PROTEIN KINASE, ... | | Authors: | Davies, T.G, Saxty, G, Woodhead, S.J, Berdini, V, Verdonk, M.L, Wyatt, P.G, Boyle, R.G, Barford, D, Downham, R, Garrett, M.D, Carr, R.A. | | Deposit date: | 2007-03-19 | | Release date: | 2007-05-08 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Identification of Inhibitors of Protein Kinase B Using Fragment-Based Lead Discovery
J.Med.Chem., 50, 2007
|
|
8HQ3
 
 | | KL1 in complex with CRM1-Ran-RanBP1 | | Descriptor: | CHLORIDE ION, CRM1 isoform 1, DIMETHYL SULFOXIDE, ... | | Authors: | Sun, Q, Jian, L. | | Deposit date: | 2022-12-13 | | Release date: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Discovery of Aminoratjadone Derivatives as Potent Noncovalent CRM1 Inhibitors.
J.Med.Chem., 66, 2023
|
|
4UR1
 
 | | Crystal structure of the PCE reductive dehalogenase from S. multivorans in complex with dibromoethene | | Descriptor: | BENZAMIDINE, CIS-DIBROMOETHENE, GLYCEROL, ... | | Authors: | Bommer, M, Kunze, C, Fesseler, J, Schubert, T, Diekert, G, Dobbek, H. | | Deposit date: | 2014-06-25 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.649 Å) | | Cite: | Structural Basis for Organohalide Respiration.
Science, 346, 2014
|
|
2UY8
 
 | | R92A mutant of Bacillus subtilis Oxalate Decarboxylase OxdC | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, MANGANESE (II) ION, OXALATE DECARBOXYLASE OXDC | | Authors: | Just, V.J, Burrell, M.R, Bowater, L, McRobbie, I, Stevenson, C.E.M, Lawson, D.M, Bornemann, S. | | Deposit date: | 2007-04-03 | | Release date: | 2007-08-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | The Identity of the Active Site of Oxalate Decarboxylase and the Importance of the Stability of Active-Site Lid Conformations.
Biochem.J., 407, 2007
|
|
8H2G
 
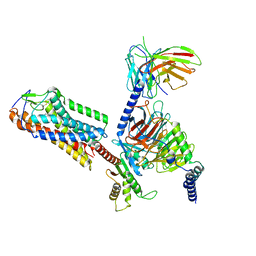 | |
4V32
 
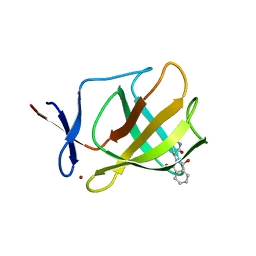 | | Cereblon isoform 4 from Magnetospirillum gryphiswaldense in complex with Thalidomide, Y101F mutant | | Descriptor: | CEREBLON ISOFORM 4, S-Thalidomide, ZINC ION | | Authors: | Hartmann, M.D, Lupas, A.N, Hernandez Alvarez, B. | | Deposit date: | 2014-10-15 | | Release date: | 2014-12-17 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Thalidomide Mimics Uridine Binding to an Aromatic Cage in Cereblon.
J.Struct.Biol., 188, 2014
|
|
2UWH
 
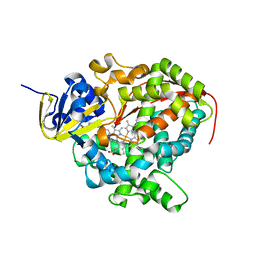 | | Cytochrome P450 BM3 mutant in complex with palmitic acid | | Descriptor: | BIFUNCTIONAL P-450: NADPH-P450 REDUCTASE, PALMITIC ACID, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Huang, W.-C, Joyce, M.G, Westlake, A.C.G, Roberts, G.C.K, Moody, P.C.E. | | Deposit date: | 2007-03-21 | | Release date: | 2007-08-28 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Filling a Hole in Cytochrome P450 Bm3 Improves Substrate Binding and Catalytic Efficiency.
J.Mol.Biol., 373, 2007
|
|
8H3A
 
 | | Cryo-EM Structure of the KBTBD2-CRL3~N8(removed)-CSN complex | | Descriptor: | COP9 signalosome complex subunit 1, COP9 signalosome complex subunit 2, COP9 signalosome complex subunit 3, ... | | Authors: | Hu, Y, Mao, Q, Chen, Z, Sun, L. | | Deposit date: | 2022-10-08 | | Release date: | 2023-10-11 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (7.51 Å) | | Cite: | Dynamic molecular architecture and substrate recruitment of cullin3-RING E3 ligase CRL3 KBTBD2.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8D9T
 
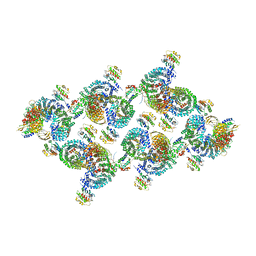 | | AP-1, Arf1, Nef lattice on MHC-I lipopeptide incorporated narrow membrane tubes, centered on gamma-Arf1 | | Descriptor: | ADP ribosylation factor 1, AP-1 complex subunit beta-1, AP-1 complex subunit gamma-1, ... | | Authors: | Hooy, R.M, Hurley, J.H. | | Deposit date: | 2022-06-11 | | Release date: | 2022-11-09 | | Last modified: | 2024-06-12 | | Method: | ELECTRON MICROSCOPY (20 Å) | | Cite: | Self-assembly and structure of a clathrin-independent AP-1:Arf1 tubular membrane coat.
Sci Adv, 8, 2022
|
|
4UXI
 
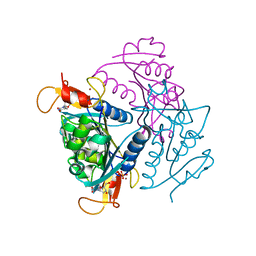 | | Leishmania major Thymidine Kinase in complex with thymidine | | Descriptor: | PHOSPHATE ION, THYMIDINE, THYMIDINE KINASE, ... | | Authors: | Timm, J, Bosch-Navarrete, C, Recio, E, Nettleship, J.E, Rada, H, Gonzalez-Pacanowska, D, Wilson, K.S. | | Deposit date: | 2014-08-22 | | Release date: | 2015-05-27 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structural and Kinetic Characterization of Thymidine Kinase from Leishmania Major.
Plos Negl Trop Dis, 9, 2015
|
|
2UWB
 
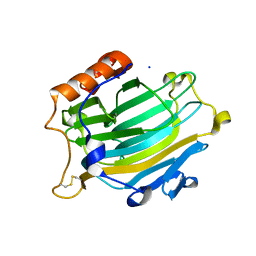 | | Crystal structure of the Nasturtium seedling mutant xyloglucanase isoform NXG1-delta-YNIIG | | Descriptor: | CELLULASE | | Authors: | Baumann, M.J, Eklof, J, Michel, G, Kallasa, A, Teeri, T.T, Brumer, H, Czjzek, M. | | Deposit date: | 2007-03-20 | | Release date: | 2007-06-26 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Evidence for the Evolution of Xyloglucanase Activity from Xyloglucan Endo-Transglycosylases: Biological Implications for Cell Wall Metabolism.
Plant Cell, 19, 2007
|
|
8H3Z
 
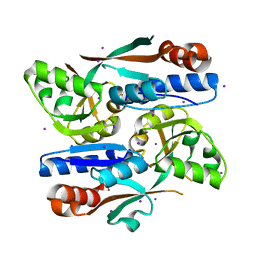 | |
4UU5
 
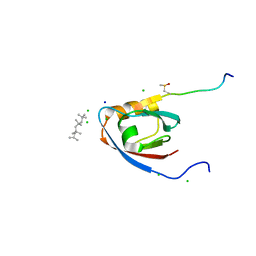 | | CRYSTAL STRUCTURE OF THE PDZ DOMAIN OF PALS1 IN COMPLEX WITH THE CRB PEPTIDE | | Descriptor: | 2,2,4,4,6,6,8-heptamethylnonane, CHLORIDE ION, MAGUK P55 SUBFAMILY MEMBER 5, ... | | Authors: | Ivanova, M.E, Purkiss, A.G, McDonald, N.Q. | | Deposit date: | 2014-07-24 | | Release date: | 2015-01-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | Structures of the human Pals1 PDZ domain with and without ligand suggest gated access of Crb to the PDZ peptide-binding groove.
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
2UY5
 
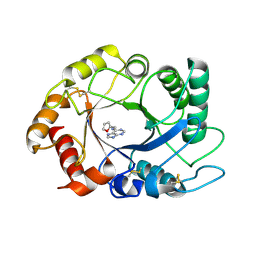 | | ScCTS1_kinetin crystal structure | | Descriptor: | ENDOCHITINASE, N-(FURAN-2-YLMETHYL)-7H-PURIN-6-AMINE | | Authors: | Hurtado-Guerrero, R, van Aalten, D.M.F. | | Deposit date: | 2007-04-02 | | Release date: | 2007-04-24 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure of Saccharomyces Cerevisiae Chitinase 1 and Screening-Based Discovery of Potent Inhibitors.
Chem.Biol., 14, 2007
|
|
