4U24
 
 | | Crystal structure of the E. coli ribosome bound to dalfopristin. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Noeske, J, Huang, J, Olivier, N.B, Giacobbe, R.A, Zambrowski, M, Cate, J.H.D. | | Deposit date: | 2014-07-16 | | Release date: | 2014-07-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Synergy of streptogramin antibiotics occurs independently of their effects on translation.
Antimicrob.Agents Chemother., 58, 2014
|
|
4U1V
 
 | | Crystal structure of the E. coli ribosome bound to linopristin. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Noeske, J, Huang, J, Olivier, N.B, Giacobbe, R.A, Zambrowski, M, Cate, J.H.D. | | Deposit date: | 2014-07-16 | | Release date: | 2014-07-30 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Synergy of streptogramin antibiotics occurs independently of their effects on translation.
Antimicrob.Agents Chemother., 58, 2014
|
|
2HU2
 
 | | CTBP/BARS in ternary complex with NAD(H) and RRTGAPPAL peptide | | Descriptor: | 9-mer peptide from Zinc finger protein 217, C-terminal-binding protein 1, FORMIC ACID, ... | | Authors: | Nardini, M, Bolognesi, M, Quinlan, K.G.R, Verger, A, Francescato, P, Crossley, M. | | Deposit date: | 2006-07-26 | | Release date: | 2006-10-31 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Specific Recognition of ZNF217 and Other Zinc Finger Proteins at a Surface Groove of C-Terminal Binding Proteins
Mol.Cell.Biol., 26, 2006
|
|
5NZ0
 
 | | Structure of Transcriptional Regulatory Repressor Protein - EthR from Mycobacterium Tuberculosis in complex with linezolid | | Descriptor: | 1,2-ETHANEDIOL, HTH-type transcriptional regulator EthR, N-{[(5S)-3-(3-fluoro-4-morpholin-4-ylphenyl)-2-oxo-1,3-oxazolidin-5-yl]methyl}acetamide | | Authors: | Blaszczyk, M, Mendes, V, Nikiforov, P.O, Blundell, T.L. | | Deposit date: | 2017-05-12 | | Release date: | 2018-05-30 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.825 Å) | | Cite: | The antibiotics linezolid and sutezolid are ligands for Mycobacterium tuberculosis EthR
To Be Published
|
|
4U26
 
 | | Crystal structure of the E. coli ribosome bound to dalfopristin and quinupristin. | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Noeske, J, Huang, J, Olivier, N.B, Giacobbe, R.A, Zambrowski, M, Cate, J.H.D. | | Deposit date: | 2014-07-16 | | Release date: | 2014-07-30 | | Last modified: | 2024-07-10 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Synergy of streptogramin antibiotics occurs independently of their effects on translation.
Antimicrob.Agents Chemother., 58, 2014
|
|
2VPL
 
 | | The structure of the complex between the first domain of L1 protein from Thermus thermophilus and mRNA from Methanococcus jannaschii | | Descriptor: | 50S RIBOSOMAL PROTEIN L1, FRAGMENT OF MRNA FOR L1-OPERON CONTAINING REGULATOR L1-BINDING SITE, POTASSIUM ION | | Authors: | Kljashtorny, V, Tishchenko, S, Nevskaya, N, Nikonov, S, Garber, M. | | Deposit date: | 2008-03-01 | | Release date: | 2008-09-23 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Domain II of Thermus thermophilus ribosomal protein L1 hinders recognition of its mRNA.
J. Mol. Biol., 383, 2008
|
|
2VQV
 
 | | Structure of HDAC4 catalytic domain with a gain-of-function mutation bound to a hydroxamic acid inhibitor | | Descriptor: | HISTONE DEACETYLASE 4, N-hydroxy-5-[(3-phenyl-5,6-dihydroimidazo[1,2-a]pyrazin-7(8H)-yl)carbonyl]thiophene-2-carboxamide, POTASSIUM ION, ... | | Authors: | Bottomley, M.J, Lo Surdo, P, Di Giovine, P, Cirillo, A, Scarpelli, R, Ferrigno, F, Jones, P, Neddermann, P, De Francesco, R, Steinkuhler, C, Gallinari, P, Carfi, A. | | Deposit date: | 2008-03-19 | | Release date: | 2008-07-22 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural and Functional Analysis of the Human Hdac4 Catalytic Domain Reveals a Regulatory Structural Zinc-Binding Domain.
J.Biol.Chem., 283, 2008
|
|
7AGH
 
 | |
7AG4
 
 | |
4DW6
 
 | | Novel N-phenyl-phenoxyacetamide derivatives as potential EthR inhibitors and ethionamide boosters. Discovery and optimization using High-Throughput Synthesis. | | Descriptor: | AMMONIUM ION, GLYCEROL, HTH-type transcriptional regulator EthR, ... | | Authors: | Flipo, M, Willand, N, Lecat-Guillet, N, Hounsou, C, Desroses, M, Leroux, F, Lens, Z, Villeret, V, Wohlkonig, A, Wintjens, R, Christophe, T, Jeon, H.K, Locht, C, Brodin, P, Baulard, A.R, Deprez, B. | | Deposit date: | 2012-02-24 | | Release date: | 2013-03-27 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of novel N-phenylphenoxyacetamide derivatives as EthR inhibitors and ethionamide boosters by combining high-throughput screening and synthesis.
J.Med.Chem., 55, 2012
|
|
7AG6
 
 | |
7AGK
 
 | | Crystal structure of E. coli SF kinase (YihV) in complex with product sulfofructose phosphate (SFP) | | Descriptor: | Sulfofructose kinase, [(2~{S},3~{S},4~{S},5~{R})-3,4,5-tris(oxidanyl)-5-(phosphonooxymethyl)oxolan-2-yl]methanesulfonic acid | | Authors: | Sharma, M, Davies, G.J. | | Deposit date: | 2020-09-22 | | Release date: | 2021-04-14 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Molecular Basis of Sulfosugar Selectivity in Sulfoglycolysis.
Acs Cent.Sci., 7, 2021
|
|
2IZN
 
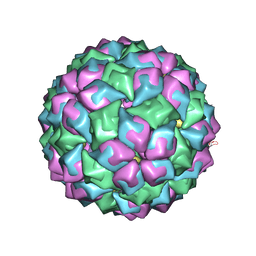 | | MS2-RNA HAIRPIN (G-10) COMPLEX | | Descriptor: | 5'-R(*AP*CP*AP*UP*CP*GP*CP*GP*AP*UP *UP*AP*CP*GP*GP*AP*UP*GP*U)-3', MS2 COAT PROTEIN | | Authors: | Helgstrand, C, Grahn, E, Moss, T, Stonehouse, N.J, Tars, K, Stockley, P.G, Liljas, L. | | Deposit date: | 2006-07-25 | | Release date: | 2006-07-27 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.56 Å) | | Cite: | Investigating the Structural Basis of Purine Specificity in the Structures of MS2 Coat Protein RNA Translational Operator Complexes
Nucleic Acids Res., 30, 2002
|
|
7JSL
 
 | |
3QH6
 
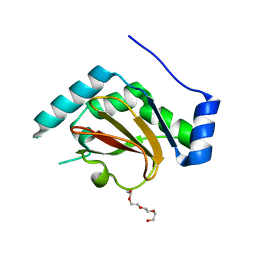 | | 1.8A resolution structure of CT296 from Chlamydia trachomatis | | Descriptor: | CT296, TETRAETHYLENE GLYCOL | | Authors: | Kemege, K, Hickey, J, Lovell, S, Battaile, K.P, Zhang, Y, Hefty, P.S. | | Deposit date: | 2011-01-25 | | Release date: | 2011-10-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ab initio structural modeling of and experimental validation for Chlamydia trachomatis protein CT296 reveal structural similarity to Fe(II) 2-oxoglutarate-dependent enzymes.
J.Bacteriol., 193, 2011
|
|
7JSA
 
 | |
1UNA
 
 | |
2P1O
 
 | | Mechanism of Auxin Perception by the TIR1 ubiquitin ligase | | Descriptor: | Auxin-responsive protein IAA7, INOSITOL HEXAKISPHOSPHATE, NAPHTHALEN-1-YL-ACETIC ACID, ... | | Authors: | Tan, X, Calderon-Villalobos, L.I.A, Sharon, M, Robinson, C.V, Estelle, M, Zheng, N. | | Deposit date: | 2007-03-06 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Mechanism of auxin perception by the TIR1 ubiquitin ligase
Nature, 446, 2007
|
|
2P1N
 
 | | Mechanism of Auxin Perception by the TIR1 Ubiqutin Ligase | | Descriptor: | (2,4-DICHLOROPHENOXY)ACETIC ACID, Auxin-responsive protein IAA7, INOSITOL HEXAKISPHOSPHATE, ... | | Authors: | Tan, X, Calderon-Villalobos, L.I.A, Sharon, M, Robinson, C.V, Estelle, M, Zheng, C, Zheng, N. | | Deposit date: | 2007-03-06 | | Release date: | 2007-04-10 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism of auxin perception by the TIR1 ubiquitin ligase
Nature, 446, 2007
|
|
8Q9R
 
 | | Crystal structure of MADS-box/MEF2D N-terminal domain bound to dsDNA and HDAC9 deacetylase binding motif | | Descriptor: | Histone deacetylase 9 (HDAC9) binding motif peptide: EVKQKLQEFLLSKS, MADS box dsDNA: AACTATTTATAAGA, MADS box dsDNA: TCTTATAAATAGT, ... | | Authors: | Chinellato, M, Carli, A, Perin, S, Mazzoccato, Y, Di Giorgio, E, Brancolini, C, Angelini, A, Cendron, L. | | Deposit date: | 2023-08-20 | | Release date: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Folding of Class IIa HDAC Derived Peptides into alpha-helices Upon Binding to Myocyte Enhancer Factor-2 in Complex with DNA.
J.Mol.Biol., 436, 2024
|
|
3B6A
 
 | | Crystal structure of the Streptomyces coelicolor TetR family protein ActR in complex with actinorhodin | | Descriptor: | 2,2'-[(1R,1'R,3S,3'S)-6,6',9,9'-tetrahydroxy-1,1'-dimethyl-5,5',10,10'-tetraoxo-3,3',4,4',5,5',10,10'-octahydro-1H,1'H-8,8'-bibenzo[g]isochromene-3,3'-diyl]diacetic acid, ActR protein | | Authors: | Willems, A.R, Junop, M.S. | | Deposit date: | 2007-10-28 | | Release date: | 2008-02-05 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Crystal structures of the Streptomyces coelicolor TetR-like protein ActR alone and in complex with actinorhodin or the actinorhodin biosynthetic precursor (S)-DNPA.
J.Mol.Biol., 376, 2008
|
|
8OIJ
 
 | | Drosophila Smaug-Smoothened complex | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Protein Smaug, ... | | Authors: | Ubartaite, G, Kubikova, J, Jeske, M. | | Deposit date: | 2023-03-23 | | Release date: | 2023-06-21 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis for binding of Drosophila Smaug to the GPCR Smoothened and to the germline inducer Oskar.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6QU1
 
 | | Crystal structure of the KAP1 RBCC domain in complex with the SMARCAD1 CUE1 domain at 3.7 angstrom resolution. | | Descriptor: | SWI/SNF-related matrix-associated actin-dependent regulator of chromatin subfamily A containing DEAD/H box 1, Transcription intermediary factor 1-beta,Transcription intermediary factor 1-beta, ZINC ION | | Authors: | Newman, J.A, Aitkenhead, H, Gavard, A, Lim, M, Williams, H.L, Svejstrup, J.Q, von Delft, F, Arrowsmith, C.H, Edwards, A, Bountra, C, Gileadi, O. | | Deposit date: | 2019-02-26 | | Release date: | 2019-07-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | A Ubiquitin-Binding Domain that Binds a Structural Fold Distinct from that of Ubiquitin.
Structure, 2019
|
|
3B6C
 
 | |
3E4U
 
 | | Crystal Structure of the Wild-Type Human BCL6 BTB/POZ Domain | | Descriptor: | B-cell lymphoma 6 protein | | Authors: | Stead, M.A, Rosbrook, G.O, Hadden, J.M, Trinh, C.H, Carr, S.B, Wright, S.C. | | Deposit date: | 2008-08-12 | | Release date: | 2008-12-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of the wild-type human BCL6 POZ domain.
Acta Crystallogr.,Sect.F, 64, 2008
|
|
