4YFL
 
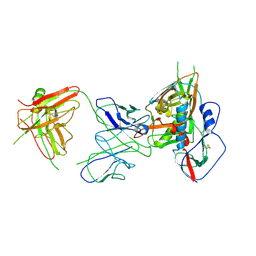 | | Crystal structure of VH1-46 germline-derived CD4-binding site-directed antibody 1B2530 in complex with HIV-1 clade A/E 93TH057 gp120 | | Descriptor: | 1B2530 Light chain, 1B2530 heavy chain, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Acharya, P, Zhou, T, Moquin, S, Kwong, P.D. | | Deposit date: | 2015-02-25 | | Release date: | 2015-06-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.387 Å) | | Cite: | Structural Repertoire of HIV-1-Neutralizing Antibodies Targeting the CD4 Supersite in 14 Donors.
Cell, 161, 2015
|
|
7YL6
 
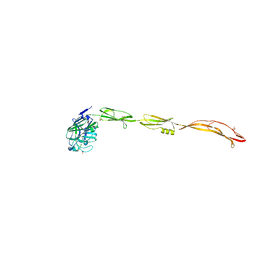 | | Cell surface protein YwfG protein complexed with alpha-1,2-mannobiose | | Descriptor: | CALCIUM ION, GRAM_POS_ANCHORING domain-containing protein, SULFATE ION, ... | | Authors: | Tsuchiya, W, Fujimoto, Z, Suzuki, C. | | Deposit date: | 2022-07-25 | | Release date: | 2023-06-07 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Cell-surface protein YwfG of Lactococcus lactis binds to alpha-1,2-linked mannose.
Plos One, 18, 2023
|
|
6H0H
 
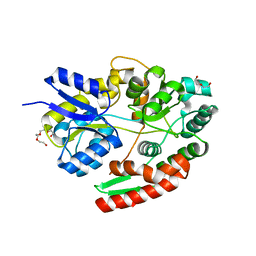 | | The ABC transporter associated binding protein from B. animalis subsp. lactis Bl-04 in complex with beta-1,6-galactobiose | | Descriptor: | DI(HYDROXYETHYL)ETHER, Probable solute binding protein of ABC transporter system for sugars, TETRAETHYLENE GLYCOL, ... | | Authors: | Fredslund, F, Lo Leggio, L. | | Deposit date: | 2018-07-09 | | Release date: | 2019-06-19 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.39 Å) | | Cite: | Substrate preference of an ABC importer corresponds to selective growth on beta-(1,6)-galactosides inBifidobacterium animalissubsp.lactis.
J.Biol.Chem., 294, 2019
|
|
6ANL
 
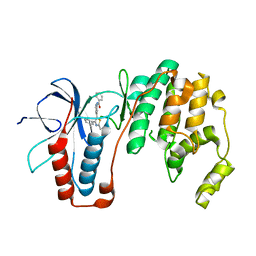 | | Structure-based Design, Synthesis, and Biological Evaluation of Imidazo[1,2-b]pyridazine-based p38 MAP Kinase Inhibitors | | Descriptor: | Mitogen-activated protein kinase 14, TAK-715 | | Authors: | Snell, G.P, Okada, K, Bragstad, K, Sang, B.-C. | | Deposit date: | 2017-08-14 | | Release date: | 2018-01-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-based design, synthesis, and biological evaluation of imidazo[1,2-b]pyridazine-based p38 MAP kinase inhibitors.
Bioorg. Med. Chem., 26, 2018
|
|
8RFG
 
 | | CgsiGP3 sample in nanodisc | | Descriptor: | Cyclic beta 1-2 glucan synthetase, beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose, beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose-(1-2)-beta-D-glucopyranose, ... | | Authors: | Sedzicki, J, Ni, D, Lehmann, F, Stahlberg, H, Dehio, C. | | Deposit date: | 2023-12-12 | | Release date: | 2024-03-20 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.35 Å) | | Cite: | Structure-function analysis of the cyclic beta-1,2-glucan synthase from Agrobacterium tumefaciens.
Nat Commun, 15, 2024
|
|
4YWG
 
 | | Crystal structure of 830A in complex with V1V2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Heavy chain of anti-HIV-1 gp120 V1V2 antibody 830A, Light chain of anti-HIV-1 gp120 V1V2 antibody 830A, ... | | Authors: | Pan, R, Kong, X. | | Deposit date: | 2015-03-20 | | Release date: | 2015-07-15 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.998 Å) | | Cite: | The V1V2 Region of HIV-1 gp120 Forms a Five-Stranded Beta Barrel.
J.Virol., 89, 2015
|
|
3MB5
 
 | |
7RAI
 
 | |
6B97
 
 | | Crystal structure of PDE2 in complex with complex 9 | | Descriptor: | 1,2-ETHANEDIOL, 6-chloro-1-methyl-N-{(1R)-1-[4-(trifluoromethyl)phenyl]ethyl}-1H-pyrazolo[3,4-d]pyrimidin-4-amine, MAGNESIUM ION, ... | | Authors: | Lu, J. | | Deposit date: | 2017-10-10 | | Release date: | 2017-11-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | The identification of a novel lead class for phosphodiesterase 2 inhibition by fragment-based drug design.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
8P67
 
 | | Crystal structure of Thermothelomyces thermophila (double mutant EE) in complex with aldotetrauronic acid | | Descriptor: | 1,2-ETHANEDIOL, 4-O-methyl-alpha-D-glucopyranuronic acid-(1-2)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose-(1-4)-beta-D-xylopyranose, GH30 family xylanase, ... | | Authors: | Dimarogona, M, Pentari, C, Kosinas, C, Topakas, E. | | Deposit date: | 2023-05-25 | | Release date: | 2024-05-22 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Structural and molecular insights into a bifunctional glycoside hydrolase 30 xylanase specific to glucuronoxylan.
Biotechnol.Bioeng., 121, 2024
|
|
6LYG
 
 | |
4CHB
 
 | | Crystal structure of the human KLHL2 Kelch domain in complex with a WNK4 peptide | | Descriptor: | 1,2-ETHANEDIOL, DODECAETHYLENE GLYCOL, KELCH-LIKE PROTEIN 2, ... | | Authors: | Sorrell, F.J, Schumacher, F.R, Kurz, T, Alessi, D.R, Newman, J, Cooper, C.D.O, Canning, P, Kopec, J, Williams, E, Krojer, T, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2013-11-29 | | Release date: | 2014-01-08 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Structural and Biochemical Characterisation of the Klhl3-Wnk Kinase Interaction Important in Blood Pressure Regulation.
Biochem.J., 460, 2014
|
|
5EM5
 
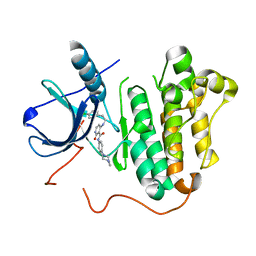 | |
1M33
 
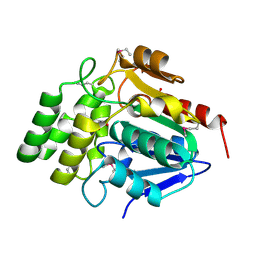 | | Crystal Structure of BioH at 1.7 A | | Descriptor: | 1,2-ETHANEDIOL, 3-HYDROXY-PROPANOIC ACID, BioH protein | | Authors: | Sanishvili, R, Savchenko, A, Skarina, T, Edwards, A, Joachimiak, A, Yakunin, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2002-06-26 | | Release date: | 2003-01-21 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Integrating structure, bioinformatics, and enzymology to discover function: BioH, a new carboxylesterase from Escherichia coli.
J.Biol.Chem., 278, 2003
|
|
8J6T
 
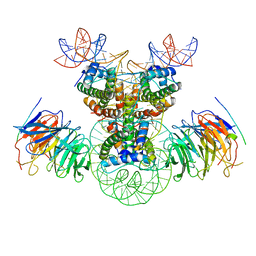 | | Cryo-EM structure of the double CAF-1 bound right-handed Di-tetrasome | | Descriptor: | Chromatin assembly factor 1 subunit A, Chromatin assembly factor 1 subunit B, Histone H3.1, ... | | Authors: | Liu, C.P, Yu, Z.Y, Xu, R.M. | | Deposit date: | 2023-04-26 | | Release date: | 2023-08-16 | | Last modified: | 2023-09-06 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Structural insights into histone binding and nucleosome assembly by chromatin assembly factor-1.
Science, 381, 2023
|
|
7DKI
 
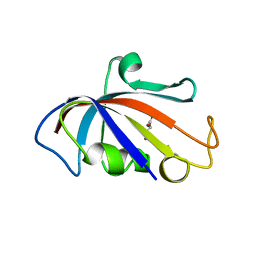 | | Silk worm FKBP, isoform-1 | | Descriptor: | GLYCEROL, Peptidylprolyl isomerase | | Authors: | Yuchi, Z, Nayak, B.C. | | Deposit date: | 2020-11-24 | | Release date: | 2022-02-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Silk worm FKBP, isoform-1
To Be Published
|
|
3MOS
 
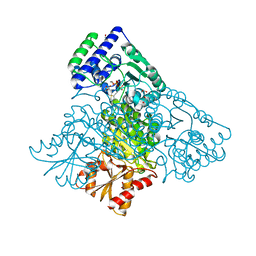 | | The structure of human Transketolase | | Descriptor: | 1,2-ETHANEDIOL, CALCIUM ION, SODIUM ION, ... | | Authors: | Parthier, C, Tittmann, K. | | Deposit date: | 2010-04-23 | | Release date: | 2010-07-28 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | The crystal structure of human transketolase and new insights into its mode of action.
J.Biol.Chem., 285, 2010
|
|
5Z1N
 
 | | Crystal structure of C terminal region of G-protein interacting protein 1 (Gip1) from Dictyostelium discoideum | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, G-protein interacting protein 1, ... | | Authors: | Miyagawa, T, Koteishi, H, Kamimura, Y, Miyanaga, Y, Takeshita, K, Nakagawa, A, Ueda, M. | | Deposit date: | 2017-12-27 | | Release date: | 2018-10-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.949 Å) | | Cite: | Structural basis of Gip1 for cytosolic sequestration of G protein in wide-range chemotaxis
Nat Commun, 9, 2018
|
|
6DHN
 
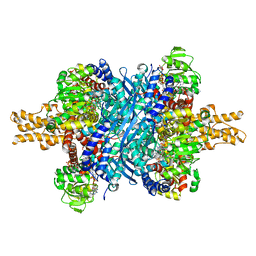 | | Bovine glutamate dehydrogenase complexed with Eu3+ | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, GLUTAMIC ACID, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Smith, T.J. | | Deposit date: | 2018-05-20 | | Release date: | 2018-07-25 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | A novel mechanism of V-type zinc inhibition of glutamate dehydrogenase results from disruption of subunit interactions necessary for efficient catalysis.
FEBS J., 278, 2011
|
|
7JFZ
 
 | |
6DIO
 
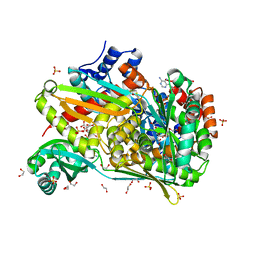 | | Structure of class II HMG-CoA reductase from Delftia acidovorans with NAD bound | | Descriptor: | 1,2-ETHANEDIOL, 3-hydroxy-3-methylglutaryl coenzyme A reductase, CITRIC ACID, ... | | Authors: | Ragwan, E.R, Arai, E, Kung, Y. | | Deposit date: | 2018-05-23 | | Release date: | 2018-09-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | New Crystallographic Snapshots of Large Domain Movements in Bacterial 3-Hydroxy-3-methylglutaryl Coenzyme A Reductase.
Biochemistry, 57, 2018
|
|
5Z39
 
 | | Crystal structure of C terminal region of G-protein interacting protein 1 (Gip1) from Dictyostelium discoideum form II | | Descriptor: | 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, DI-PALMITOYL-3-SN-PHOSPHATIDYLETHANOLAMINE, G-protein interacting protein 1, ... | | Authors: | Miyagawa, T, Koteishi, H, Kamimura, Y, Miyanaga, Y, Takeshita, K, Nakagawa, A, Ueda, M. | | Deposit date: | 2018-01-05 | | Release date: | 2018-10-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structural basis of Gip1 for cytosolic sequestration of G protein in wide-range chemotaxis
Nat Commun, 9, 2018
|
|
5EIJ
 
 | | Carbonic Anhydrase II in complex with Sulfonamide Inhibitor | | Descriptor: | 1-(3-iodanylphenyl)-3-(4-sulfamoylphenyl)thiourea, Carbonic anhydrase 2, DIMETHYL SULFOXIDE, ... | | Authors: | Lomelino, C.L, Mahon, B.P, McKenna, R. | | Deposit date: | 2015-10-29 | | Release date: | 2016-11-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Kinetic and X-ray crystallographic investigations on carbonic anhydrase isoforms I, II, IX and XII of a thioureido analog of SLC-0111.
Bioorg. Med. Chem., 24, 2016
|
|
5K36
 
 | | Structure of an eleven component nuclear RNA exosome complex bound to RNA | | Descriptor: | Exosome complex component CSL4, Exosome complex component MTR3, Exosome complex component RRP4, ... | | Authors: | Lima, C.D, Zinder, J.C. | | Deposit date: | 2016-05-19 | | Release date: | 2016-11-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Nuclear RNA Exosome at 3.1 angstrom Reveals Substrate Specificities, RNA Paths, and Allosteric Inhibition of Rrp44/Dis3.
Mol.Cell, 64, 2016
|
|
6U2K
 
 | |
