1BLA
 
 | | BASIC FIBROBLAST GROWTH FACTOR (FGF-2) MUTANT WITH CYS 78 REPLACED BY SER AND CYS 96 REPLACED BY SER, NMR | | Descriptor: | BASIC FIBROBLAST GROWTH FACTOR | | Authors: | Powers, R, Seddon, A.P, Bohlen, P, Moy, F.J. | | Deposit date: | 1996-05-20 | | Release date: | 1996-11-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution structure of basic fibroblast growth factor determined by multidimensional heteronuclear magnetic resonance spectroscopy.
Biochemistry, 35, 1996
|
|
2MJR
 
 | | Anoplin R5W structure in DPC micelles | | Descriptor: | Anoplin, UNKNOWN ATOM OR ION, dodecyl 2-(trimethylammonio)ethyl phosphate | | Authors: | Uggerhoej, L, Poulsen, T.J, Wimmer, R. | | Deposit date: | 2014-01-16 | | Release date: | 2014-12-03 | | Last modified: | 2024-11-20 | | Method: | SOLUTION NMR | | Cite: | Rational Design of Alpha-Helical Antimicrobial Peptides: Do's and Don'ts.
Chembiochem, 16, 2015
|
|
2MA4
 
 | | Solution NMR Structure of yahO protein from Salmonella typhimurium, Northeast Structural Genomics Consortium (NESG) Target StR106 | | Descriptor: | Putative periplasmic protein | | Authors: | Eletsky, A, Zhang, Q, Liu, G, Wang, H, Nwosu, C, Cunningham, K, Ma, L, Xiao, R, Liu, J, Baran, M.C, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2013-06-27 | | Release date: | 2013-08-28 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structural and Functional Characterization of DUF1471 Domains of Salmonella Proteins SrfN, YdgH/SssB, and YahO.
Plos One, 9, 2014
|
|
2Z3I
 
 | | Crystal structure of blasticidin S deaminase (BSD) mutant E56Q complexed with substrate | | Descriptor: | BLASTICIDIN S, Blasticidin-S deaminase, CACODYLATE ION, ... | | Authors: | Kumasaka, T, Yamamoto, M, Furuichi, M, Nakasako, M, Kimura, M, Yamaguchi, I, Ueki, T. | | Deposit date: | 2007-06-04 | | Release date: | 2007-10-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of blasticidin S deaminase (BSD): implications for dynamic properties of catalytic zinc
J.Biol.Chem., 282, 2007
|
|
2MJQ
 
 | |
2Z3H
 
 | | Crystal structure of blasticidin S deaminase (BSD) complexed with deaminohydroxy blasticidin S | | Descriptor: | 1-(4-{[(3R)-3-AMINO-5-{[(Z)-AMINO(IMINO)METHYL](METHYL)AMINO}PENTANOYL]AMINO}-2,3,4-TRIDEOXY-D-ERYTHRO-HEX-2-ENOPYRANURONOSYL)-4-HYDROXYPYRIMIDIN-2(1H)-ONE, Blasticidin-S deaminase, ZINC ION | | Authors: | Kumasaka, T, Yamamoto, M, Furuichi, M, Nakasako, M, Kimura, M, Yamaguchi, I, Ueki, T. | | Deposit date: | 2007-06-04 | | Release date: | 2007-10-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of blasticidin S deaminase (BSD): implications for dynamic properties of catalytic zinc
J.Biol.Chem., 282, 2007
|
|
2MUS
 
 | | HADDOCK calculated model of LIN5001 bound to the HET-s amyloid | | Descriptor: | 3''',4'-bis(carboxymethyl)-2,2':5',2'':5'',2''':5''',2''''-quinquethiophene-5,5''''-dicarboxylic acid, Heterokaryon incompatibility protein s | | Authors: | Hermann, U.S, Schuetz, A.K, Shirani, H, Saban, D, Nuvolone, M, Huang, D.H, Li, B, Ballmer, B, Aslund, A.K.O, Mason, J.J, Rushing, E, Budka, H, Hammarstrom, P, Bockmann, A, Caflisch, A, Meier, B.H, Nilsson, P.K.R, Hornemann, S, Aguzzi, A. | | Deposit date: | 2014-09-16 | | Release date: | 2017-02-01 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structure-based drug design identifies polythiophenes as antiprion compounds.
Sci Transl Med, 7, 2015
|
|
2Z3G
 
 | | Crystal structure of blasticidin S deaminase (BSD) | | Descriptor: | Blasticidin-S deaminase, ZINC ION, alpha-D-glucopyranose-(1-1)-alpha-D-glucopyranose | | Authors: | Kumasaka, T, Yamamoto, M, Furuichi, M, Nakasako, M, Kimura, M, Yamaguchi, I, Ueki, T. | | Deposit date: | 2007-06-04 | | Release date: | 2007-10-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structures of blasticidin S deaminase (BSD): implications for dynamic properties of catalytic zinc
J.Biol.Chem., 282, 2007
|
|
1WBA
 
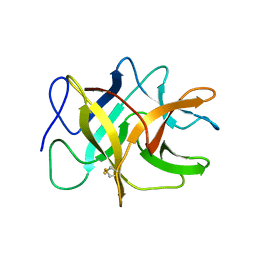 | | WINGED BEAN ALBUMIN 1 | | Descriptor: | WINGED BEAN ALBUMIN 1 | | Authors: | Mccoy, A.J. | | Deposit date: | 1996-06-19 | | Release date: | 1997-01-11 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystallization and preliminary crystallographic data of the major albumin from Psophocarpus tetragonolobus (L.) DC.
J.Biol.Chem., 262, 1987
|
|
2IOG
 
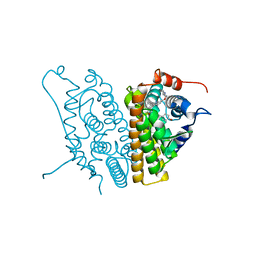 | |
2IOK
 
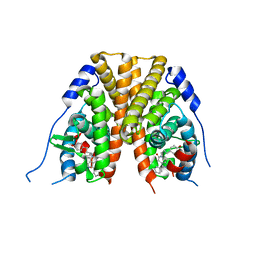 | |
1WAV
 
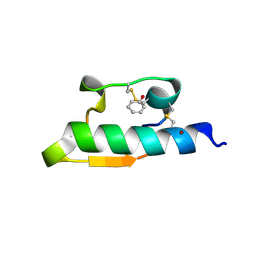 | | CRYSTAL STRUCTURE OF FORM B MONOCLINIC CRYSTAL OF INSULIN | | Descriptor: | INSULIN, PHENOL, ZINC ION | | Authors: | Liang, D.-C, Ding, J.-H, Chang, W.-R, Wan, Z.-L. | | Deposit date: | 1996-02-28 | | Release date: | 1997-02-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular replacement study on form-B monoclinic crystal of insulin.
Sci.China, Ser.C: Life Sci., 39, 1996
|
|
2HLD
 
 | | Crystal structure of yeast mitochondrial F1-ATPase | | Descriptor: | ATP synthase alpha chain, mitochondrial, ATP synthase beta chain, ... | | Authors: | Kabaleeswaran, V, Puri, N, Walker, J.E, Leslie, A.G, Mueller, D.M. | | Deposit date: | 2006-07-06 | | Release date: | 2006-11-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Novel features of the rotary catalytic mechanism revealed in the structure of yeast F(1) ATPase.
Embo J., 25, 2006
|
|
2PQL
 
 | | Crystal Structure of Anopheles gambiae D7R4-tryptamine complex | | Descriptor: | 2-(1H-INDOL-3-YL)ETHANAMINE, D7r4 protein | | Authors: | Andersen, J.F, Mans, B.J, Calvo, E, Ribeiro, J.M. | | Deposit date: | 2007-05-02 | | Release date: | 2007-10-09 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Crystal Structure of D7r4, a Salivary Biogenic Amine-binding Protein from the Malaria Mosquito Anopheles gambiae.
J.Biol.Chem., 282, 2007
|
|
1WKY
 
 | | Crystal structure of alkaline mannanase from Bacillus sp. strain JAMB-602: catalytic domain and its Carbohydrate Binding Module | | Descriptor: | CALCIUM ION, CHLORIDE ION, SODIUM ION, ... | | Authors: | Akita, M, Takeda, N, Hirasawa, K, Sakai, H, Kawamoto, M, Yamamoto, M, Grant, W.D, Hatada, Y, Ito, S, Horikoshi, K. | | Deposit date: | 2004-06-15 | | Release date: | 2005-06-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystallization and preliminary X-ray study of alkaline mannanase from an alkaliphilic Bacillus isolate.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1WQ2
 
 | | Neutron Crystal Structure Of Dissimilatory Sulfite Reductase D (DsrD) | | Descriptor: | Protein dsvD, SULFATE ION | | Authors: | Chatake, T, Mizuno, N, Voordouw, G, Higuchi, Y, Arai, S, Tanaka, I, Niimura, N. | | Deposit date: | 2004-09-19 | | Release date: | 2005-09-19 | | Last modified: | 2023-10-25 | | Method: | NEUTRON DIFFRACTION (2.4 Å) | | Cite: | Crystallization and preliminary neutron analysis of the dissimilatory sulfite reductase D (DsrD) protein from the sulfate-reducing bacterium Desulfovibrio vulgaris.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
2Z3J
 
 | | Crystal structure of blasticidin S deaminase (BSD) R90K mutant | | Descriptor: | Blasticidin-S deaminase, CACODYLATE ION, CHLORIDE ION, ... | | Authors: | Teh, A.H, Kumasaka, T, Yamamoto, M, Furuichi, M, Nakasako, M, Kimura, M, Yamaguchi, I, Ueki, T. | | Deposit date: | 2007-06-04 | | Release date: | 2007-10-23 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structures of blasticidin S deaminase (BSD): implications for dynamic properties of catalytic zinc
J.Biol.Chem., 282, 2007
|
|
2Q0I
 
 | | Structure of Pseudomonas Quinolone Signal Response Protein PqsE | | Descriptor: | BENZOIC ACID, FE (III) ION, Quinolone signal response protein | | Authors: | Yu, S, Jensen, V, Feldmann, I, Haussler, S, Blankenfeldt, W. | | Deposit date: | 2007-05-22 | | Release date: | 2008-06-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structure elucidation and preliminary assessment of hydrolase activity of PqsE, the Pseudomonas quinolone signal (PQS) response protein.
Biochemistry, 48, 2009
|
|
1V75
 
 | | Crystal structure of hemoglobin D from the Aldabra giant tortoise (Geochelone gigantea) at 2.0 A resolution | | Descriptor: | Hemoglobin A and D beta chain, Hemoglobin D alpha chain, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Kuwada, T, Hasegawa, T, Satoh, I, Ishikawa, K, Shishikura, F. | | Deposit date: | 2003-12-12 | | Release date: | 2003-12-30 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Crystallization and preliminary X-ray diffraction study of hemoglobin D from the Aldabra giant tortoise, Geochelone gigantea.
Protein Pept.Lett., 10, 2003
|
|
1BLD
 
 | | BASIC FIBROBLAST GROWTH FACTOR (FGF-2) MUTANT WITH CYS 78 REPLACED BY SER AND CYS 96 REPLACED BY SER, NMR | | Descriptor: | BASIC FIBROBLAST GROWTH FACTOR | | Authors: | Powers, R, Seddon, A.P, Bohlen, P, Moy, F.J. | | Deposit date: | 1996-05-20 | | Release date: | 1996-11-08 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | High-resolution solution structure of basic fibroblast growth factor determined by multidimensional heteronuclear magnetic resonance spectroscopy.
Biochemistry, 35, 1996
|
|
1VBK
 
 | | Crystal structure of PH1313 from Pyrococcus horikoshii Ot3 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, hypothetical protein PH1313 | | Authors: | Sugahara, M, Kunishima, N, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2004-02-27 | | Release date: | 2005-04-19 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Purification, crystallization and preliminary crystallographic analysis of the putative thiamine-biosynthesis protein PH1313 from Pyrococcus horikoshii OT3
Acta Crystallogr.,Sect.F, 63, 2007
|
|
1EWJ
 
 | | CRYSTAL STRUCTURE OF BLEOMYCIN-BINDING PROTEIN COMPLEXED WITH BLEOMYCIN | | Descriptor: | BLEOMYCIN A2, BLEOMYCIN RESISTANCE DETERMINANT | | Authors: | Maruyama, M, Kumagai, T, Matoba, Y, Hata, Y, Sugiyama, M. | | Deposit date: | 2000-04-26 | | Release date: | 2001-04-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structures of the transposon Tn5-carried bleomycin resistance determinant uncomplexed and complexed with bleomycin.
J.Biol.Chem., 276, 2001
|
|
2Q0J
 
 | | Structure of Pseudomonas Quinolone Signal Response Protein PqsE | | Descriptor: | BENZOIC ACID, FE (III) ION, Quinolone signal response protein | | Authors: | Yu, S, Jensen, V, Feldmann, I, Haussler, S, Blankenfeldt, W. | | Deposit date: | 2007-05-22 | | Release date: | 2008-06-03 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure elucidation and preliminary assessment of hydrolase activity of PqsE, the Pseudomonas quinolone signal (PQS) response protein.
Biochemistry, 48, 2009
|
|
2G1Q
 
 | | crystal structure of KSP in complex with inhibitor 9h | | Descriptor: | (5S)-5-(3-AMINOPROPYL)-3-(2,5-DIFLUOROPHENYL)-N-ETHYL-5-PHENYL-4,5-DIHYDRO-1H-PYRAZOLE-1-CARBOXAMIDE, ADENOSINE-5'-DIPHOSPHATE, Kinesin-like protein KIF11, ... | | Authors: | Yan, Y. | | Deposit date: | 2006-02-14 | | Release date: | 2006-10-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Kinesin spindle protein (KSP) inhibitors. Part 4: Structure-based design of 5-alkylamino-3,5-diaryl-4,5-dihydropyrazoles as potent, water-soluble inhibitors of the mitotic kinesin KSP.
Bioorg.Med.Chem.Lett., 16, 2006
|
|
1ECS
 
 | | THE 1.7 A CRYSTAL STRUCTURE OF A BLEOMYCIN RESISTANCE DETERMINANT ENCODED ON THE TRANSPOSON TN5 | | Descriptor: | BLEOMYCIN RESISTANCE PROTEIN, CALCIUM ION, TETRAETHYLENE GLYCOL | | Authors: | Maruyama, M, Matoba, Y, Kumagai, T, Sugiyama, M. | | Deposit date: | 2000-01-25 | | Release date: | 2001-05-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structures of the transposon Tn5-carried bleomycin resistance determinant uncomplexed and complexed with bleomycin.
J.Biol.Chem., 276, 2001
|
|
