3NG2
 
 | |
5N9T
 
 | | Crystal structure of USP7 in complex with a potent, selective and reversible small-molecule inhibitor | | Descriptor: | 3-[4-(aminomethyl)phenyl]-2-methyl-6-[[4-oxidanyl-1-[(3~{R})-4,4,4-tris(fluoranyl)-3-phenyl-butanoyl]piperidin-4-yl]methyl]pyrazolo[4,3-d]pyrimidin-7-one, GLYCEROL, SULFATE ION, ... | | Authors: | Harrison, T, Gavory, G, O'Dowd, C, Helm, M, Flasz, J, Arkoudis, E, Dossang, A, Hughes, C, Cassidy, E, McClelland, K, Odrzywol, E, Page, N, Barker, O, Miel, H. | | Deposit date: | 2017-02-27 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.73 Å) | | Cite: | Discovery and characterization of highly potent and selective allosteric USP7 inhibitors.
Nat. Chem. Biol., 14, 2018
|
|
3NGQ
 
 | | Crystal structure of the human CNOT6L nuclease domain | | Descriptor: | 3-PYRIDINIUM-1-YLPROPANE-1-SULFONATE, CCR4-NOT transcription complex subunit 6-like, MAGNESIUM ION | | Authors: | Wang, H, Morita, M, Yang, W, Bartlam, M, Yamamoto, T, Rao, Z. | | Deposit date: | 2010-06-13 | | Release date: | 2010-07-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of the human CNOT6L nuclease domain reveals strict poly(A) substrate specificity.
Embo J., 2010
|
|
3HW8
 
 | | ternary complex of DNA polymerase lambda of a two nucleotide gapped DNA substrate with a C in the scrunch site | | Descriptor: | 1,2-ETHANEDIOL, 2',3'-DIDEOXY-THYMIDINE-5'-TRIPHOSPHATE, 5'-D(*CP*AP*GP*TP*AP*T)-3', ... | | Authors: | Garcia-Diaz, M, Bebenek, K, Larrea, A.A, Havener, J.M, Perera, L, Krahn, J.M, Pedersen, L.C, Ramsden, D.A, Kunkel, T.A. | | Deposit date: | 2009-06-17 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Scrunching Durning DNA Repair Synthesis
To be Published
|
|
2V3F
 
 | | acid-beta-glucosidase produced in carrot | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLUCOSYLCERAMIDASE, SULFATE ION, ... | | Authors: | Shaaltiel, Y, Bartfeld, D, Hashmueli, S, Baum, G, Brill-Almon, E, Galili, G, Dym, O, Boldin-Adamsky, S.A, Silman, I, Sussman, J.L, Futerman, A.H, Aviezer, D, Israel Structural Proteomics Center (ISPC) | | Deposit date: | 2007-06-17 | | Release date: | 2008-04-08 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Production of Glucocerebrosidase with Terminal Mannose Glycans for Enzyme Replacement Therapy of Gaucher'S Disease Using a Plant Cell System.
Plant Biotechnol.J., 5, 2007
|
|
3HWT
 
 | | Ternary complex of DNA polymerase lambda bound to a two nucleotide gapped DNA substrate with a scrunched dA | | Descriptor: | 1,2-ETHANEDIOL, 2',3'-DIDEOXY-THYMIDINE-5'-TRIPHOSPHATE, 5'-D(*CP*AP*GP*TP*AP*(2DT))-3', ... | | Authors: | Garcia-Diaz, M, Bebenek, K, Larrea, A.A, Pedersen, L.C, Kunkel, T.A. | | Deposit date: | 2009-06-18 | | Release date: | 2009-08-25 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Scrunching during DNA repair synthesis
To be Published
|
|
5O10
 
 | | Y48H mutant of human cytochrome c | | Descriptor: | Cytochrome c, HEME C | | Authors: | Moreno-Chicano, T, Deacon, O.M, Hough, M.A, Worrall, J.A.R. | | Deposit date: | 2017-05-17 | | Release date: | 2018-03-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Heightened Dynamics of the Oxidized Y48H Variant of Human Cytochrome c Increases Its Peroxidatic Activity.
Biochemistry, 56, 2017
|
|
1KKA
 
 | |
3I57
 
 | | Type 2 repeat of the mucus binding protein MUB from Lactobacillus reuteri | | Descriptor: | CALCIUM ION, Mucus binding protein | | Authors: | Hemmings, A.M, MacKenzie, D.A, Tailford, L.E, Juge, N. | | Deposit date: | 2009-07-03 | | Release date: | 2009-09-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of a mucus binding protein repeat reveals an unexpected functional immunoglobulin binding activity.
J.Biol.Chem., 284, 2009
|
|
2WEK
 
 | | Crystal structure of the human MGC45594 gene product in complex with diclofenac | | Descriptor: | 2-[2,6-DICHLOROPHENYL)AMINO]BENZENEACETIC ACID, CHLORIDE ION, GLYCEROL, ... | | Authors: | Shafqat, N, Yue, W.W, Ugochukwu, E, Niesen, F, Smee, C, Arrowsmith, C, Weigelt, J, Edwards, A, Bountra, C, Oppermann, U. | | Deposit date: | 2009-03-31 | | Release date: | 2009-12-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal Structure of the Human Mgc45594 Gene Product in Complex with Diclofenac
To be Published
|
|
3NGN
 
 | | Crystal structure of the human CNOT6L nuclease domain in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, CCR4-NOT transcription complex subunit 6-like | | Authors: | Wang, H, Morita, M, Yang, W, Bartlam, M, Yamamoto, T, Rao, Z. | | Deposit date: | 2010-06-12 | | Release date: | 2010-07-28 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure of the human CNOT6L nuclease domain reveals strict poly(A) substrate specificity.
Embo J., 2010
|
|
2WCG
 
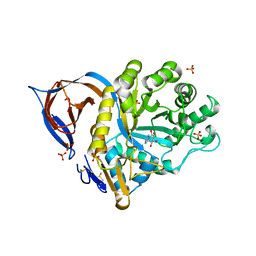 | | X-ray structure of acid-beta-glucosidase with N-octyl(cyclic guanidine)-nojirimycin in the active site | | Descriptor: | CHLORIDE ION, GLUCOSYLCERAMIDASE, N-[(3E,5R,6R,7S,8R,8AR)-5,6,7,8-TETRAHYDROXYHEXAHYDROIMIDAZO[1,5-A]PYRIDIN-3(2H)-YLIDENE]OCTAN-1-AMINIUM, ... | | Authors: | Brumshtein, B, Aguilar, M, Garcia-Moreno, M.I, Mellet, C.O, Garcia-Fernandez, J.M, Silman, I, Shaaltiel, Y, Aviezer, D, Sussman, J.L, Futerman, A.H. | | Deposit date: | 2009-03-12 | | Release date: | 2009-11-24 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | 6-Amino-6-Deoxy-5,6-Di-N-(N'-Octyliminomethylidene)Nojirimycin: Synthesis, Biological Evaluation, and Crystal Structure in Complex with Acid Beta-Glucosidase.
Chembiochem, 10, 2009
|
|
3N5U
 
 | | Crystal structure of an Rb C-terminal peptide bound to the catalytic subunit of PP1 | | Descriptor: | CHLORIDE ION, MANGANESE (II) ION, Retinoblastoma-associated protein, ... | | Authors: | Hirschi, A.M, Cecchini, M, Steinhardt, R.C, Dick, F.A, Rubin, S.M. | | Deposit date: | 2010-05-25 | | Release date: | 2010-08-11 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | An overlapping kinase and phosphatase docking site regulates activity of the retinoblastoma protein.
Nat.Struct.Mol.Biol., 17, 2010
|
|
5NTU
 
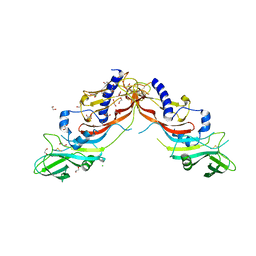 | | Crystal Structure of human Pro-myostatin Precursor at 2.6 A Resolution | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Growth/differentiation factor 8 | | Authors: | Cotton, T.R, Fischer, G, Hyvonen, M. | | Deposit date: | 2017-04-28 | | Release date: | 2018-01-17 | | Last modified: | 2018-02-21 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structure of the human myostatin precursor and determinants of growth factor latency.
EMBO J., 37, 2018
|
|
5NUB
 
 | |
2WFT
 
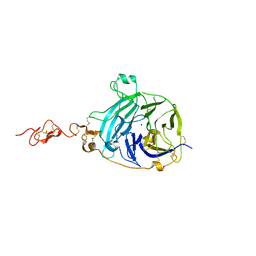 | | Crystal structure of the human HIP ectodomain | | Descriptor: | CHLORIDE ION, HEDGEHOG-INTERACTING PROTEIN, SODIUM ION | | Authors: | Bishop, B, Aricescu, A.R, Harlos, K, O'Callaghan, C.A, Jones, E.Y, Siebold, C. | | Deposit date: | 2009-04-15 | | Release date: | 2009-06-30 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural Insights Into Hedgehog Ligand Sequestration by the Human Hedgehog-Interacting Protein Hip
Nat.Struct.Mol.Biol., 16, 2009
|
|
5NVM
 
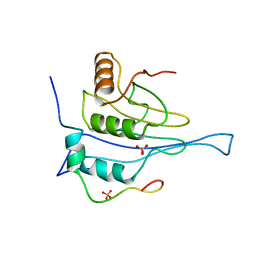 | |
3I9J
 
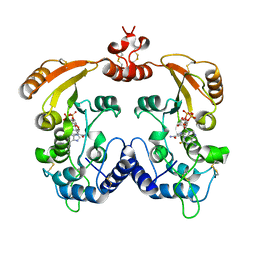 | | Crystal structure of ADP ribosyl cyclase complexed with a substrate analog and a product nicotinamide | | Descriptor: | ADP-ribosyl cyclase, NICOTINAMIDE, Nicotinamide 2-fluoro-adenine dinucleotide, ... | | Authors: | Liu, Q, Graeff, R, Kriksunov, I.A, Jiang, H, Zhang, B, Oppenheimer, N, Lin, H, Potter, B.V.L, Lee, H.C, Hao, Q. | | Deposit date: | 2009-07-12 | | Release date: | 2009-07-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural basis for enzymatic evolution from a dedicated ADP-ribosyl cyclase to a multifunctional NAD hydrolase
J.Biol.Chem., 284, 2009
|
|
5O6Z
 
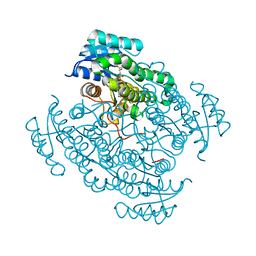 | | 17beta-hydroxysteroid dehydrogenase 14 variant T205 in complex with a non-steroidal quinoline based inhibitor | | Descriptor: | (4-fluoranyl-3-oxidanyl-phenyl)-quinolin-2-yl-methanone, 17-beta-hydroxysteroid dehydrogenase 14, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Bertoletti, N, Braun, F, Heine, A, Klebe, G, Marchais-Oberwinkler, S. | | Deposit date: | 2017-06-07 | | Release date: | 2018-06-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Structure-based design and profiling of novel 17 beta-HSD14 inhibitors.
Eur J Med Chem, 155, 2018
|
|
3NA1
 
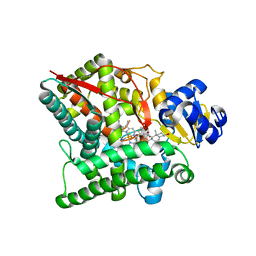 | | Crystal structure of human CYP11A1 in complex with 20-hydroxycholesterol | | Descriptor: | (3alpha,8alpha)-cholest-5-ene-3,20-diol, Adrenodoxin, mitochondrial, ... | | Authors: | Strushkevich, N.V, MacKenzie, F, Tempel, W, Botchkarev, A, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Weigelt, J.U, Park, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-05-31 | | Release date: | 2011-06-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural basis for pregnenolone biosynthesis by the mitochondrial monooxygenase system.
Proc.Natl.Acad.Sci.USA, 108, 2011
|
|
3I8C
 
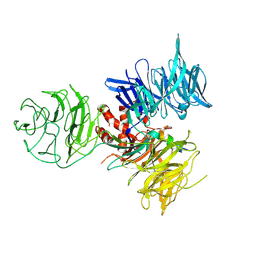 | | Crystal Structure of DDB1 in Complex with the H-Box Motif of WDR21A | | Descriptor: | DNA damage-binding protein 1, WD repeat-containing protein 21A | | Authors: | Li, T, Robert, E.I, Breugel, P.C.V, Strubin, M, Zheng, N. | | Deposit date: | 2009-07-09 | | Release date: | 2009-12-08 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A promiscuous alpha-helical motif anchors viral hijackers and substrate receptors to the CUL4-DDB1 ubiquitin ligase machinery.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3I9L
 
 | | Crystal structure of ADP ribosyl cyclase complexed with N1-cIDPR | | Descriptor: | ADP-ribosyl cyclase, N1-CYCLIC INOSINE 5'-DIPHOSPHORIBOSE | | Authors: | Liu, Q, Graeff, R, Kriksunov, I.A, Jiang, H, Zhang, B, Oppenheimer, N, Lin, H, Potter, B.V.L, Lee, H.C, Hao, Q. | | Deposit date: | 2009-07-12 | | Release date: | 2009-07-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for enzymatic evolution from a dedicated ADP-ribosyl cyclase to a multifunctional NAD hydrolase
J.Biol.Chem., 284, 2009
|
|
1K0O
 
 | | Crystal structure of a soluble form of CLIC1. An intracellular chloride ion channel | | Descriptor: | CHLORIDE INTRACELLULAR CHANNEL PROTEIN 1 | | Authors: | Harrop, S.J, DeMaere, M.Z, Fairlie, W.D, Reztsova, T, Valenzuela, S.M, Mazzanti, M, Tonini, R, Qiu, M.R, Jankova, L, Warton, K, Bauskin, A.R, Wu, W.M, Pankhurst, S, Campbell, T.J, Breit, S.N, Curmi, P.M.G. | | Deposit date: | 2001-09-19 | | Release date: | 2001-12-12 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of a soluble form of the intracellular chloride ion channel CLIC1 (NCC27) at 1.4-A resolution.
J.Biol.Chem., 276, 2001
|
|
3N4C
 
 | | 6-Phenyl-1H-imidazo[4,5-c]pyridine-4-carbonitrile as cathepsin S inhibitors | | Descriptor: | (E)-1-(1-methyl-6-{4-[3-(4-methylpiperazin-1-yl)propoxy]-3-(trifluoromethyl)phenyl}-1H-imidazo[4,5-c]pyridin-4-yl)methanimine, Cathepsin S, DIMETHYL SULFOXIDE, ... | | Authors: | Fradera, X, Uitdehaag, J.C.M, van Zeeland, M. | | Deposit date: | 2010-05-21 | | Release date: | 2011-04-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | 6-Phenyl-1H-imidazo[4,5-c]pyridine-4-carbonitrile as cathepsin S inhibitors.
Bioorg.Med.Chem.Lett., 20, 2010
|
|
5NU7
 
 | | Structure of human holo plasma RBP4 | | Descriptor: | CHLORIDE ION, RETINOL, Retinol-binding protein 4 | | Authors: | Perduca, M, Monaco, H.L, Galliano, M. | | Deposit date: | 2017-04-28 | | Release date: | 2018-03-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Human plasma retinol-binding protein (RBP4) is also a fatty acid-binding protein.
Biochim. Biophys. Acta, 1863, 2018
|
|
