8USK
 
 | |
6ZPM
 
 | | Crystal structure of the unconventional kinetochore protein Trypanosoma cruzi KKT4 coiled coil domain | | Descriptor: | THREONINE, Trypanosoma cruzi KKT4 117-218 | | Authors: | Ludzia, P, Lowe, D.E, Marciano, G, Mohammed, S, Redfield, C, Akiyoshi, B. | | Deposit date: | 2020-07-08 | | Release date: | 2020-10-21 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural characterization of KKT4, an unconventional microtubule-binding kinetochore protein.
Structure, 29, 2021
|
|
7T3H
 
 | | MicroED structure of Dynobactin | | Descriptor: | TRP-ASN-SER-ASN-VAL-HIS-SER-TYR-ARG-PHE | | Authors: | Yoo, B.-K, Kaiser, J.T, Rees, D.C, Miller, R.D, Iinishi, A, Lewis, K, Bowman, S. | | Deposit date: | 2021-12-07 | | Release date: | 2022-10-19 | | Method: | ELECTRON CRYSTALLOGRAPHY (1.05 Å) | | Cite: | Computational identification of a systemic antibiotic for gram-negative bacteria.
Nat Microbiol, 7, 2022
|
|
8USE
 
 | |
8QD6
 
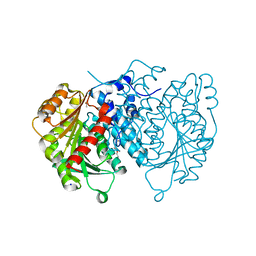 | | AntI Ser245DHA (PMSF) | | Descriptor: | ACETATE ION, Photorhabdus luminescens subsp. laumondii TTO1 complete genome segment 15/17, SODIUM ION, ... | | Authors: | Schmalhofer, M, Vagstad, A.L, Zhou, Q, Bode, H.B, Groll, M. | | Deposit date: | 2023-08-28 | | Release date: | 2024-03-13 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Polyketide Trimming Shapes Dihydroxynaphthalene-Melanin and Anthraquinone Pigments.
Adv Sci, 11, 2024
|
|
6N54
 
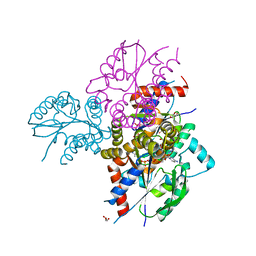 | | Crystal structure of human uridine-cytidine kinase 2 complexed with 2'-azidocytidine monophosphate | | Descriptor: | 2'-azido-2'-deoxycytidine 5'-(dihydrogen phosphate), GLYCEROL, MAGNESIUM ION, ... | | Authors: | Cuthbert, B.J, Nainar, S, Spitale, R.C, Goulding, C.W. | | Deposit date: | 2018-11-21 | | Release date: | 2019-11-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.424 Å) | | Cite: | An optimized chemical-genetic method for cell-specific metabolic labeling of RNA.
Nat.Methods, 17, 2020
|
|
6X66
 
 | | Legionella pneumophila dDot T4SS OMC | | Descriptor: | DotC, DotD, Inner membrane lipoprotein YiaD, ... | | Authors: | Durie, C.L, Sheedlo, M.J, Chung, J.M, Byrne, B.G, Su, M, Knight, T, Swanson, M.S, Lacy, D.B, Ohi, M.D. | | Deposit date: | 2020-05-27 | | Release date: | 2020-10-28 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural analysis of the Legionella pneumophila Dot/Icm type IV secretion system core complex.
Elife, 9, 2020
|
|
7TOR
 
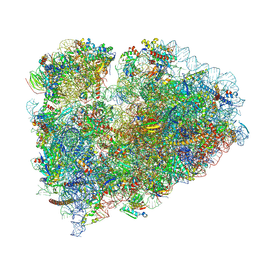 | | Mammalian 80S ribosome bound with the ALS/FTD-associated dipeptide repeat protein GR20 | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Svidritskiy, E, Susorov, D, Lee, S, Park, A, Zvornicanin, S, Demo, G, Gao, F.B, Korostelev, A.A. | | Deposit date: | 2022-01-24 | | Release date: | 2022-05-25 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Ribosome inhibition by C9ORF72-ALS/FTD-associated poly-PR and poly-GR proteins revealed by cryo-EM.
Nat Commun, 13, 2022
|
|
8USL
 
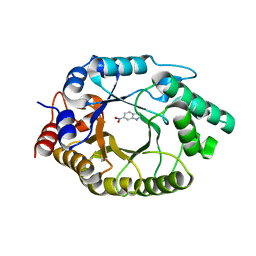 | |
8QN3
 
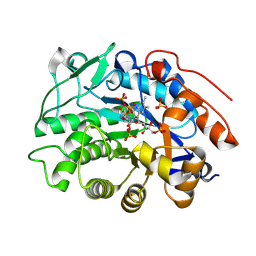 | | OPR3 wildtype in complex with NADH4 | | Descriptor: | 1,4,5,6-Tetrahydronicotinamide adenine dinucleotide, 12-oxophytodienoate reductase 3, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Bijelic, A, Macheroux, P, Keschbaumer, B. | | Deposit date: | 2023-09-25 | | Release date: | 2024-01-17 | | Last modified: | 2024-04-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Loop 6 and the beta-hairpin flap are structural hotspots that determine cofactor specificity in the FMN-dependent family of ene-reductases.
Febs J., 291, 2024
|
|
8USJ
 
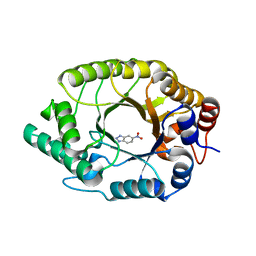 | |
4Y4L
 
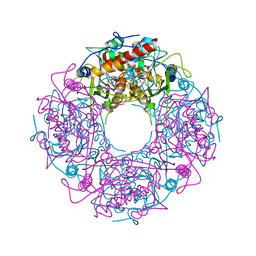 | | Crystal structure of yeast Thi4-C205S | | Descriptor: | (2E)-2-[(2S,4R)-5-[[[(2R,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]methoxy-oxidanyl-phosphoryl]oxy-oxidanyl-phosphoryl]oxy-4-oxidanyl-3-oxidanylidene-pentan-2-yl]iminoethanoic acid, Thiamine thiazole synthase | | Authors: | Zhang, X, Ealick, S.E. | | Deposit date: | 2015-02-10 | | Release date: | 2016-03-09 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Basis for Iron-Mediated Sulfur Transfer in Archael and Yeast Thiazole Synthases.
Biochemistry, 55, 2016
|
|
8C76
 
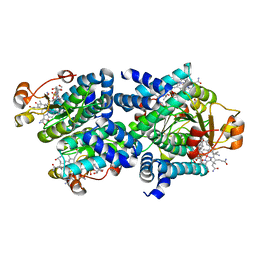 | | Light-state 2.5 Angstrom wild-type X-ray crystal structure of the cobalamin binding domain belonging to a light-dependent transcription regulator TtCarH obtained under aerobic conditions from a form 2 crystal illuminated during 5 s | | Descriptor: | COBALAMIN, Probable transcriptional regulator | | Authors: | Rios-Santacruz, R, Colletier, J.P, Schiro, G, Weik, M. | | Deposit date: | 2023-01-12 | | Release date: | 2023-08-09 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Redox driven B 12 -ligand switch drives CarH photoresponse.
Nat Commun, 14, 2023
|
|
8USG
 
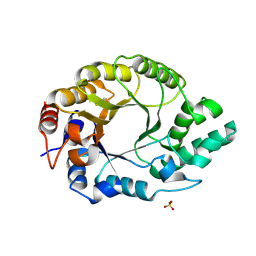 | |
7TOQ
 
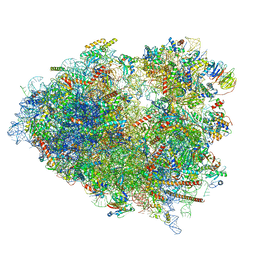 | | Mammalian 80S ribosome bound with the ALS/FTD-associated dipeptide repeat protein poly-PR | | Descriptor: | 18S rRNA, 28S rRNA, 40S ribosomal protein S10, ... | | Authors: | Loveland, A.B, Svidritskiy, E, Susorov, D, Lee, S, Park, A, Zvornicanin, S, Demo, G, Gao, F.B, Korostelev, A.A. | | Deposit date: | 2022-01-24 | | Release date: | 2022-05-25 | | Last modified: | 2024-02-21 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Ribosome inhibition by C9ORF72-ALS/FTD-associated poly-PR and poly-GR proteins revealed by cryo-EM.
Nat Commun, 13, 2022
|
|
5DZV
 
 | | Protocadherin alpha 7 extracellular cadherin domains 1-5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Protein Pcdha7, ... | | Authors: | Goodman, K.M, Bahna, F, Honig, B, Shapiro, L. | | Deposit date: | 2015-09-26 | | Release date: | 2016-05-04 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Structural Basis of Diverse Homophilic Recognition by Clustered alpha- and beta-Protocadherins.
Neuron, 90, 2016
|
|
6DNU
 
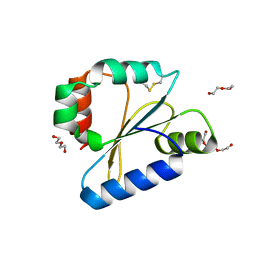 | |
6XPP
 
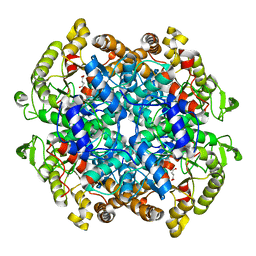 | |
4Y85
 
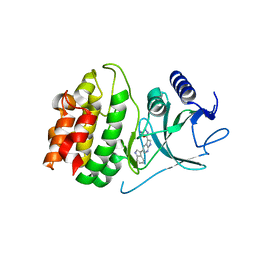 | | Crystal structure of COT kinase domain in complex with 5-(5-(1H-indol-3-yl)-1H-pyrrolo[2,3-b]pyridin-3-yl)-1,3,4-oxadiazol-2-amine | | Descriptor: | 5-[5-(1H-indol-3-yl)-1H-pyrrolo[2,3-b]pyridin-3-yl]-1,3,4-oxadiazol-2-amine, Mitogen-activated protein kinase kinase kinase 8 | | Authors: | Gutmann, S, Hinniger, A. | | Deposit date: | 2015-02-16 | | Release date: | 2015-05-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | The Crystal Structure of Cancer Osaka Thyroid Kinase Reveals an Unexpected Kinase Domain Fold.
J.Biol.Chem., 290, 2015
|
|
5IZV
 
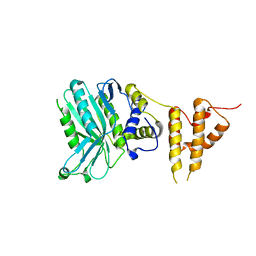 | | Crystal structure of the legionella pneumophila effector protein RavZ - F222 | | Descriptor: | Uncharacterized protein RavZ | | Authors: | Kwon, D.H, Kim, L, Kim, B.-W, Hong, S.B, Song, H.K. | | Deposit date: | 2016-03-26 | | Release date: | 2016-11-09 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.814 Å) | | Cite: | The 1:2 complex between RavZ and LC3 reveals a mechanism for deconjugation of LC3 on the phagophore membrane
Autophagy, 13, 2017
|
|
7NYJ
 
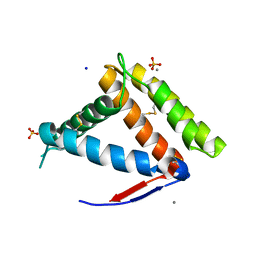 | | Structure of OBP1 from Varroa destructor, form P3<2>21 | | Descriptor: | CALCIUM ION, Odorant Binding Protein 1 from Varoa destructor, form P3<2>21, ... | | Authors: | Cambillau, C, Amigues, B, Roussel, A, Leone, P, Gaubert, A, Pelosi, P. | | Deposit date: | 2021-03-22 | | Release date: | 2021-07-07 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | A new non-classical fold of varroa odorant-binding proteins reveals a wide open internal cavity.
Sci Rep, 11, 2021
|
|
7TOO
 
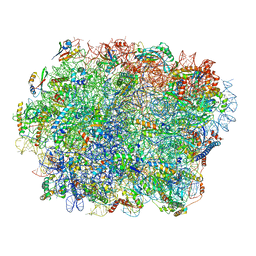 | | Yeast 80S ribosome bound with the ALS/FTD-associated dipeptide repeat protein GR20 | | Descriptor: | 25S rRNA, 5.8S rRNA, 5S rRNA, ... | | Authors: | Loveland, A.B, Svidritskiy, E, Susorov, D, Lee, S, Park, A, Zvornicanin, S, Demo, G, Gao, F.B, Korostelev, A.A. | | Deposit date: | 2022-01-24 | | Release date: | 2022-05-25 | | Last modified: | 2022-06-01 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Ribosome inhibition by C9ORF72-ALS/FTD-associated poly-PR and poly-GR proteins revealed by cryo-EM.
Nat Commun, 13, 2022
|
|
8C36
 
 | |
5E63
 
 | | K262A mutant of I-SmaMI | | Descriptor: | 2-(2-METHOXYETHOXY)ETHANOL, 2-METHOXYETHANOL, DNA (5'-D(P*CP*AP*GP*GP*TP*GP*TP*AP*CP*G)-3'), ... | | Authors: | Shen, B, Stoddard, B. | | Deposit date: | 2015-10-09 | | Release date: | 2016-01-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Structural Basis of Asymmetry in DNA Binding and Cleavage as Exhibited by the I-SmaMI LAGLIDADG Meganuclease.
J.Mol.Biol., 428, 2016
|
|
6WBS
 
 | | Human CFTR first nucleotide binding domain with dF508/V510D | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Cystic fibrosis transmembrane conductance regulator, MAGNESIUM ION | | Authors: | Simon, K.S, Kothe, M, Hilbert, B, Batchelor, J.D, Hurlbut, G.D. | | Deposit date: | 2020-03-27 | | Release date: | 2021-04-07 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.857 Å) | | Cite: | Determining the Molecular Mechanism of Suppressor Mutation V510D and the Contribution of Helical Unraveling to the dF508-CFTR Defect
To Be Published
|
|
