4GP5
 
 | | Structure of Recombinant Cytochrome ba3 Oxidase mutant Y133W from Thermus thermophilus | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, COPPER (II) ION, Cytochrome c oxidase polypeptide 2A, ... | | Authors: | Li, Y, Chen, Y, Stout, C.D. | | Deposit date: | 2012-08-20 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Ligand Access to the Active Site in Thermus thermophilusba(3) and Bovine Heart aa(3) Cytochrome Oxidases.
Biochemistry, 52, 2013
|
|
6MT9
 
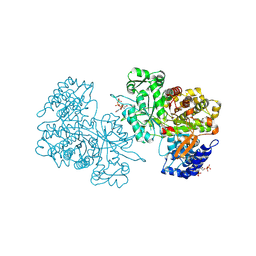 | | X-ray crystal structure of Bacillus subtilis ribonucleotide reductase NrdE alpha subunit with TTP, ATP, and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Thomas, W.C, Brooks, F.P, Bacik, J.P, Ando, N. | | Deposit date: | 2018-10-19 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Convergent allostery in ribonucleotide reductase.
Nat Commun, 10, 2019
|
|
6MMH
 
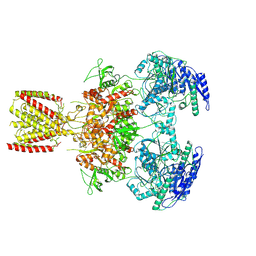 | | Diheteromeric NMDA receptor GluN1/GluN2A in the 'Extended-2' conformation, in complex with glycine and glutamate, in the presence of 1 millimolar zinc chloride, and at pH 7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, NMDA 1, ... | | Authors: | Jalali-Yazdi, F, Chowdhury, S, Yoshioka, C, Gouaux, E. | | Deposit date: | 2018-09-30 | | Release date: | 2018-11-28 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (8.21 Å) | | Cite: | Mechanisms for Zinc and Proton Inhibition of the GluN1/GluN2A NMDA Receptor.
Cell, 175, 2018
|
|
6MMW
 
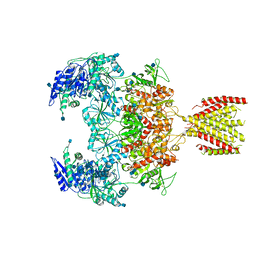 | | Triheteromeric NMDA receptor GluN1/GluN2A/GluN2A* in the '2-Knuckle-Symmetric' conformation, in complex with glycine and glutamate, in the presence of 1 micromolar zinc chloride, and at pH 7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | Authors: | Jalali-Yazdi, F, Chowdhury, S, Yoshioka, C, Gouaux, E. | | Deposit date: | 2018-10-01 | | Release date: | 2018-11-28 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (6.2 Å) | | Cite: | Mechanisms for Zinc and Proton Inhibition of the GluN1/GluN2A NMDA Receptor.
Cell, 175, 2018
|
|
6MUY
 
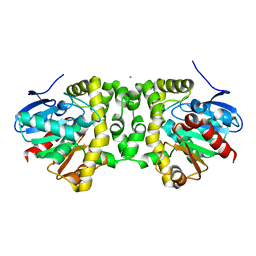 | | Fluoroacetate dehalogenase, room temperature structure solved by serial 3 degree oscillation crystallography | | Descriptor: | CALCIUM ION, Fluoroacetate dehalogenase | | Authors: | Finke, A.D, Wierman, J.L, Pare-Labrosse, O, Sarrachini, A, Besaw, J, Mehrabi, P, Gruner, S.M, Miller, R.J.D. | | Deposit date: | 2018-10-24 | | Release date: | 2019-03-27 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Fixed-target serial oscillation crystallography at room temperature.
IUCrJ, 6, 2019
|
|
4GTR
 
 | | FTase in complex with BMS analogue 13 | | Descriptor: | 4-({(3R)-7-cyano-4-[(4-methoxyphenyl)sulfonyl]-1-[(1-methyl-1H-imidazol-5-yl)methyl]-2,3,4,5-tetrahydro-1H-1,4-benzodiazepin-3-yl}methyl)phenyl diethylcarbamate, DIMETHYL SULFOXIDE, FARNESYL DIPHOSPHATE, ... | | Authors: | Guo, Z, Stigter, E.A, Bon, R.S, Waldmann, H, Blankenfeldt, W, Goody, R.S. | | Deposit date: | 2012-08-29 | | Release date: | 2012-10-24 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Development of Selective, Potent RabGGTase Inhibitors
J.Med.Chem., 55, 2012
|
|
6MV9
 
 | | X-ray crystal structure of Bacillus subtilis ribonucleotide reductase NrdE alpha subunit with TTP and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, Ribonucleoside-diphosphate reductase, ... | | Authors: | Thomas, W.C, Brooks, F.P, Bacik, J.P, Ando, N. | | Deposit date: | 2018-10-24 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | Convergent allostery in ribonucleotide reductase.
Nat Commun, 10, 2019
|
|
4LND
 
 | | Crystal structure of human apurinic/apyrimidinic endonuclease 1 with essential Mg2+ cofactor | | Descriptor: | DNA-(apurinic or apyrimidinic site) lyase, MAGNESIUM ION | | Authors: | Manvilla, B.A, Pozharski, E, Toth, E.A, Drohat, A.C. | | Deposit date: | 2013-07-11 | | Release date: | 2013-11-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure of human apurinic/apyrimidinic endonuclease 1 with the essential Mg(2+) cofactor.
Acta Crystallogr.,Sect.D, 69, 2013
|
|
4GWK
 
 | | Crystal structure of 6-phosphogluconate dehydrogenase complexed with 3-phosphoglyceric acid | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 3-PHOSPHOGLYCERIC ACID, 6-phosphogluconate dehydrogenase, ... | | Authors: | He, C, Zhou, L, Zhang, L. | | Deposit date: | 2012-09-03 | | Release date: | 2012-11-28 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.534 Å) | | Cite: | Phosphoglycerate mutase 1 coordinates glycolysis and biosynthesis to promote tumor growth.
Cancer Cell, 22, 2012
|
|
7AHI
 
 | | Substrate-engaged type 3 secretion system needle complex from Salmonella enterica typhimurium - SpaR state 2 | | Descriptor: | 1,2-DIACYL-GLYCEROL-3-SN-PHOSPHATE, LAURYL DIMETHYLAMINE-N-OXIDE, Lipoprotein PrgK, ... | | Authors: | Fahrenkamp, D, Goessweiner-Mohr, N, Miletic, S, Wald, J, Marlovits, T. | | Deposit date: | 2020-09-24 | | Release date: | 2021-03-17 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Substrate-engaged type III secretion system structures reveal gating mechanism for unfolded protein translocation
Nat Commun, 12, 2021
|
|
6MK4
 
 | |
7AGX
 
 | | Apo-state type 3 secretion system export apparatus complex from Salmonella enterica typhimurium | | Descriptor: | Protein PrgI, Protein PrgJ, Surface presentation of antigens protein SpaP, ... | | Authors: | Goessweiner-Mohr, N, Fahrenkamp, D, Miletic, S, Wald, J, Marlovits, T. | | Deposit date: | 2020-09-23 | | Release date: | 2021-03-17 | | Last modified: | 2024-05-01 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Substrate-engaged type III secretion system structures reveal gating mechanism for unfolded protein translocation.
Nat Commun, 12, 2021
|
|
7K4M
 
 | | Crystal structure of MetAP2 Modified Hemoglobin S | | Descriptor: | CARBON MONOXIDE, Hemoglobin subunit alpha, Hemoglobin subunit beta, ... | | Authors: | Musayev, F.N, Safo, M.K, Light, D.R. | | Deposit date: | 2020-09-15 | | Release date: | 2021-10-13 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | MetAP2 inhibition modifies hemoglobin S to delay polymerization and improves blood flow in sickle cell disease.
Blood Adv, 5, 2021
|
|
7B0N
 
 | |
4LML
 
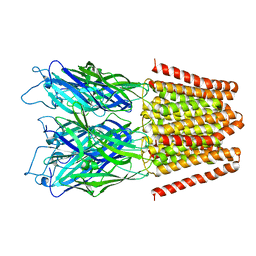 | | GLIC double mutant I9'A T25'A | | Descriptor: | Proton-gated ion channel | | Authors: | Grosman, C, Gonzalez-Gutierrez, G. | | Deposit date: | 2013-07-10 | | Release date: | 2013-10-30 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Gating of the proton-gated ion channel from Gloeobacter violaceus at pH 4 as revealed by X-ray crystallography.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
4HAG
 
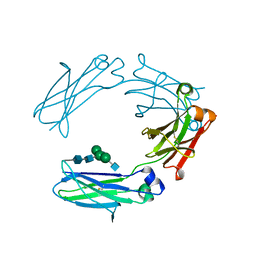 | | Crystal structure of fc-fragment of human IgG2 antibody (centered crystal form) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Ig gamma-2 chain C region | | Authors: | Teplyakov, A, Malia, T, Obmolova, G, Zhao, Y, Gilliland, G. | | Deposit date: | 2012-09-26 | | Release date: | 2013-06-12 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | IgG2 Fc structure and the dynamic features of the IgG CH2-CH3 interface.
Mol.Immunol., 56, 2013
|
|
4HFI
 
 | | The GLIC pentameric Ligand-Gated Ion Channel at 2.4 A resolution | | Descriptor: | ACETATE ION, CHLORIDE ION, DIUNDECYL PHOSPHATIDYL CHOLINE, ... | | Authors: | Sauguet, L, Corringer, P.J, Delarue, M. | | Deposit date: | 2012-10-05 | | Release date: | 2013-02-20 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for ion permeation mechanism in pentameric ligand-gated ion channels.
Embo J., 32, 2013
|
|
4LOU
 
 | |
6MKO
 
 | |
3I19
 
 | |
6MMA
 
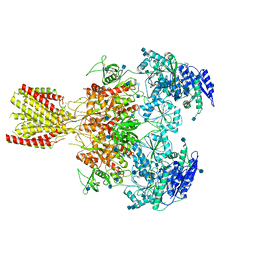 | | Diheteromeric NMDA receptor GluN1/GluN2A in the 'Extended' conformation, in complex with glycine and glutamate, in the presence of 1 micromolar zinc chloride, and at pH 6.1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, NMDA 1, ... | | Authors: | Jalali-Yazdi, F, Chowdhury, S, Yoshioka, C, Gouaux, E. | | Deposit date: | 2018-09-29 | | Release date: | 2018-11-28 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (6.31 Å) | | Cite: | Mechanisms for Zinc and Proton Inhibition of the GluN1/GluN2A NMDA Receptor.
Cell, 175, 2018
|
|
3I1J
 
 | | Structure of a putative short chain dehydrogenase from Pseudomonas syringae | | Descriptor: | 1,2-ETHANEDIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Singer, A.U, Evdokimova, E, Kudritska, M, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-06-26 | | Release date: | 2009-07-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of a putative short chain dehydrogenase from Pseudomonas syringae
To be Published
|
|
6MMT
 
 | | Triheteromeric NMDA receptor GluN1/GluN2A/GluN2A* in the '1-Knuckle' conformation, in complex with glycine and glutamate, in the presence of 1 micromolar zinc chloride, and at pH 7.4 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Glutamate receptor ionotropic, ... | | Authors: | Jalali-Yazdi, F, Chowdhury, S, Yoshioka, C, Gouaux, E. | | Deposit date: | 2018-10-01 | | Release date: | 2018-11-28 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (7.46 Å) | | Cite: | Mechanisms for Zinc and Proton Inhibition of the GluN1/GluN2A NMDA Receptor.
Cell, 175, 2018
|
|
3I2H
 
 | | Cocaine Esterase with mutation L169K, bound to DTT adduct | | Descriptor: | (4S,5S)-4,5-BIS(MERCAPTOMETHYL)-1,3-DIOXOLAN-2-OL, CHLORIDE ION, Cocaine esterase, ... | | Authors: | Tesmer, J.J.G, Nance, M.R. | | Deposit date: | 2009-06-29 | | Release date: | 2010-06-16 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Structural analysis of thermostabilizing mutations of cocaine esterase.
Protein Eng.Des.Sel., 23, 2010
|
|
7JNI
 
 | | Crystal structure of the angiotensin II type 2 receptoror (AT2R) in complex with EMA401 | | Descriptor: | (2R)-2,3-dihydroxypropyl (9Z)-octadec-9-enoate, FORMIC ACID, HEXANE-1,6-DIOL, ... | | Authors: | Cherezov, V, Shaye, H, Han, G.W. | | Deposit date: | 2020-08-04 | | Release date: | 2022-02-09 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Inhibition of the angiotensin II type 2 receptor AT 2 R is a novel therapeutic strategy for glioblastoma.
Proc.Natl.Acad.Sci.USA, 119, 2022
|
|
