8EZW
 
 | |
6UST
 
 | | Gut microbial sulfatase from Hungatella hathewayi | | Descriptor: | CALCIUM ION, N-acetylgalactosamine 6-sulfate sulfatase | | Authors: | Ervin, S.M, Redinbo, M.R. | | Deposit date: | 2019-10-28 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Insights into Endobiotic Reactivation by Human Gut Microbiome-Encoded Sulfatases.
Biochemistry, 59, 2020
|
|
8EZ2
 
 | | Plasmodium falciparum M1 in complex with inhibitor 15ag | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, M1 family aminopeptidase, ... | | Authors: | Calic, P.P.S, McGowan, S, Webb, C.T. | | Deposit date: | 2022-10-31 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structure-based development of potent Plasmodium falciparum M1 and M17 aminopeptidase selective and dual inhibitors via S1'-region optimisation.
Eur.J.Med.Chem., 248, 2022
|
|
4ZOG
 
 | |
4ZOP
 
 | | Co-crystal Structure of Lipid Kinase PI3K alpha with a selective phosphatidylinositol-3 kinase alpha inhibitor | | Descriptor: | (2S,3R)-N~1~-(8-tert-butyl-4,5-dihydro[1,3]thiazolo[4,5-h]quinazolin-2-yl)-3-methylpyrrolidine-1,2-dicarboxamide, Phosphatidylinositol 3-kinase regulatory subunit alpha, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit alpha isoform | | Authors: | Knapp, M.S, Elling, R.A. | | Deposit date: | 2015-05-06 | | Release date: | 2016-05-18 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | Co-crystal Structure of the Lipid Kinase PI3K alpha with a selective phosphatidylinositol-3 kinase alpha inhibitorCo-crystal Structure of the Lipid Kinase PI3K alpha with a selective phosphatidylinositol-3 kinase alpha inhibitor
To Be Published
|
|
8HP5
 
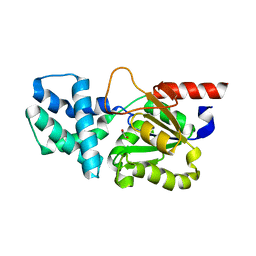 | | Crystal structure of (S)-2-haloacid dehalogenase | | Descriptor: | (S)-2-haloacid dehalogenase, 1,2-ETHANEDIOL | | Authors: | Yang, Q, Wang, L, Xu, X, Xing, X, Zhou, J. | | Deposit date: | 2022-12-12 | | Release date: | 2023-06-21 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Enzymatic hydrolysis on L-azetidine-2-carboxylate ring opening
Catalysis Science And Technology, 2023
|
|
4ZRF
 
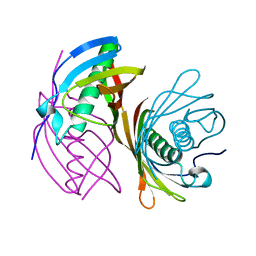 | |
8EYF
 
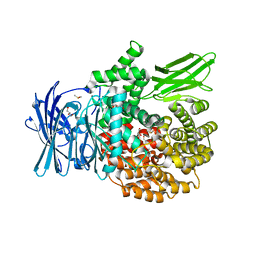 | | Plasmodium falciparum M1 in complex with inhibitor 15aa | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, M1 family aminopeptidase, ... | | Authors: | Calic, P.P.S, McGowan, S, Webb, C.T. | | Deposit date: | 2022-10-26 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure-based development of potent Plasmodium falciparum M1 and M17 aminopeptidase selective and dual inhibitors via S1'-region optimisation.
Eur.J.Med.Chem., 248, 2022
|
|
5JXE
 
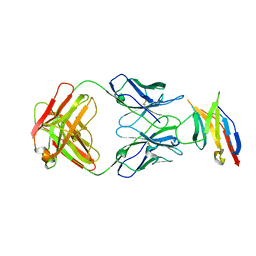 | | Human PD-1 ectodomain complexed with Pembrolizumab Fab | | Descriptor: | Pembrolizumab Fab heavy chain, Pembrolizumab Fab light chain, Programmed cell death protein 1 | | Authors: | Na, Z, Bharath, S.R, Song, H. | | Deposit date: | 2016-05-13 | | Release date: | 2016-08-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis for blocking PD-1-mediated immune suppression by therapeutic antibody pembrolizumab.
Cell Res., 27, 2017
|
|
6UW3
 
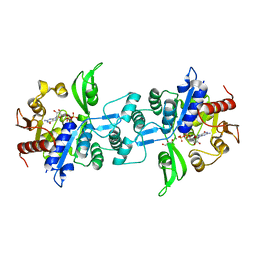 | | The crystal structure of FbiA from Mycobacterium Smegmatis, GDP Bound form | | Descriptor: | CALCIUM ION, GLYCEROL, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Grinter, R, Gillett, D, Cordero, P.R.F, Greening, C. | | Deposit date: | 2019-11-04 | | Release date: | 2020-05-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Cellular and Structural Basis of Synthesis of the Unique Intermediate Dehydro-F420-0 in Mycobacteria.
mSystems, 5, 2020
|
|
8EZ4
 
 | | Plasmodium falciparum M17 in complex with inhibitor 9aa | | Descriptor: | CARBONATE ION, M17 leucyl aminopeptidase, N-[(1R)-2-(hydroxyamino)-2-oxo-1-(3',4',5'-trifluoro[1,1'-biphenyl]-4-yl)ethyl]-N~2~-phenylglycinamide, ... | | Authors: | Calic, P.P.S, McGowan, S, Webb, C.T. | | Deposit date: | 2022-10-31 | | Release date: | 2023-01-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structure-based development of potent Plasmodium falciparum M1 and M17 aminopeptidase selective and dual inhibitors via S1'-region optimisation.
Eur.J.Med.Chem., 248, 2022
|
|
8GUV
 
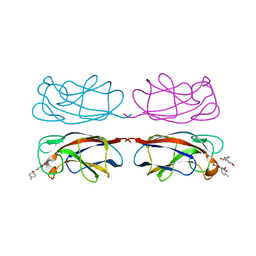 | | LecA from Pseudomonas aeruginosa in complex with tolcapone (CAS: 134308-13-7) | | Descriptor: | CALCIUM ION, PA-I galactophilic lectin, Tolcapone | | Authors: | Kuhaudomlarp, S, Siebs, E, Varrot, A, Imberty, A, Titz, A. | | Deposit date: | 2022-09-13 | | Release date: | 2023-07-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | LecA from Pseudomonas aeruginosa in complex with tolcapone (CAS: 134308-13-7)
To Be Published
|
|
5JYI
 
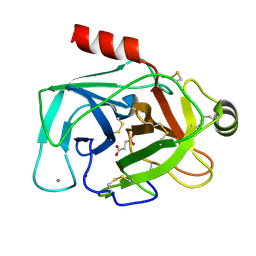 | | Trypsin bound with succinic acid at 1.9A | | Descriptor: | CALCIUM ION, Cationic trypsin, SODIUM ION, ... | | Authors: | Manohar, R, Kutumbarao, N.H.V, KarthiK, L, Malathy, P, Velmurugan, D, Gunasekaran, K. | | Deposit date: | 2016-05-14 | | Release date: | 2016-07-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.914 Å) | | Cite: | Trypsin bound with succinic acid at 1.9A
To Be Published
|
|
5JZK
 
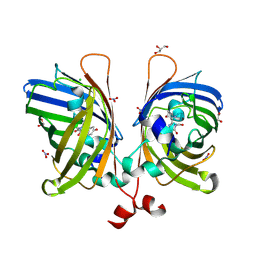 | | The Structure of Ultra Stable Green Fluorescent Protein | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Yong, K.J, Gunn, N.J, Scott, D.J, Griffin, M.D.W. | | Deposit date: | 2016-05-17 | | Release date: | 2017-12-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Novel Ultra-Stable, Monomeric Green Fluorescent Protein For Direct Volumetric Imaging of Whole Organs Using CLARITY.
Sci Rep, 8, 2018
|
|
6UWO
 
 | |
6UWU
 
 | | Crystal Structure of the first bromodomain of human BRD4 in complex with the inhibitor ZL0516 | | Descriptor: | 1,2-ETHANEDIOL, 2-{4-[(2R)-2-hydroxy-3-(4-methylpiperazin-1-yl)propoxy]-3,5-dimethylphenyl}-5,7-dimethoxy-4H-1-benzopyran-4-one, Bromodomain-containing protein 4 | | Authors: | Leonard, P.G, Joseph, S. | | Deposit date: | 2019-11-05 | | Release date: | 2020-04-15 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of Orally Bioavailable Chromone Derivatives as Potent and Selective BRD4 Inhibitors: Scaffold Hopping, Optimization, and Pharmacological Evaluation.
J.Med.Chem., 63, 2020
|
|
5JZB
 
 | | Crystal structure of HsaD bound to 3,5-dichlorobenzene sulphonamide | | Descriptor: | 3,5-dichlorobenzene-1-sulfonamide, 4,5:9,10-diseco-3-hydroxy-5,9,17-trioxoandrosta-1(10),2-diene-4-oate hydrolase, PHOSPHATE ION | | Authors: | Ryan, A, Polycarpou, E, Lack, N.A, Evangelopoulos, D, Sieg, C, Halman, A, Bhakta, S, Sinclair, A, Eleftheriadou, O, McHugh, T.D, Keany, S, Lowe, E, Ballet, R, Abihammad, A, Ciulli, A, Sim, E. | | Deposit date: | 2016-05-16 | | Release date: | 2017-04-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.102 Å) | | Cite: | Investigation of the mycobacterial enzyme HsaD as a potential novel target for anti-tubercular agents using a fragment-based drug design approach.
Br. J. Pharmacol., 174, 2017
|
|
5JZN
 
 | | Crystal structure of DCLK1-KD in complex with NVP-TAE684 | | Descriptor: | 5-CHLORO-N-[2-METHOXY-4-[4-(4-METHYLPIPERAZIN-1-YL)PIPERIDIN-1-YL]PHENYL]-N'-(2-PROPAN-2-YLSULFONYLPHENYL)PYRIMIDINE-2,4-DIAMINE, SULFATE ION, Serine/threonine-protein kinase DCLK1 | | Authors: | Patel, O, Lucet, I. | | Deposit date: | 2016-05-17 | | Release date: | 2016-08-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Biochemical and Structural Insights into Doublecortin-like Kinase Domain 1.
Structure, 24, 2016
|
|
6UY3
 
 | |
4Z09
 
 | | Crystal structure of FVO strain Plasmodium falciparum AMA1 in complex with the RON2hp [Thr2040Ala] peptide | | Descriptor: | Apical membrane antigen 1, GLYCEROL, Rhoptry neck protein 2 | | Authors: | Wang, G, McGowan, S, Norton, R.S, Scanlon, M.J. | | Deposit date: | 2015-03-26 | | Release date: | 2016-08-03 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structure-Activity Studies of beta-Hairpin Peptide Inhibitors of the Plasmodium falciparum AMA1-RON2 Interaction.
J.Mol.Biol., 428, 2016
|
|
5K4D
 
 | | Structure of eukaryotic translation initiation factor 3 subunit D (eIF3d) cap binding domain from Nasonia vitripennis, Crystal form 3 | | Descriptor: | Eukaryotic translation initiation factor 3 subunit D | | Authors: | Kranzusch, P.J, Lee, A.S.Y, Doudna, J.A, Cate, J.H.D. | | Deposit date: | 2016-05-20 | | Release date: | 2016-07-27 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | eIF3d is an mRNA cap-binding protein that is required for specialized translation initiation.
Nature, 536, 2016
|
|
5K4X
 
 | | M. thermoresistible IMPDH in complex with IMP and Compound 1 | | Descriptor: | INOSINIC ACID, Inosine-5'-monophosphate dehydrogenase,Inosine-5'-monophosphate dehydrogenase, ~{N}-(2~{H}-indazol-6-yl)-3,5-dimethyl-1~{H}-pyrazole-4-sulfonamide | | Authors: | Pacitto, A, Ascher, D.B, Blundell, T.L. | | Deposit date: | 2016-05-22 | | Release date: | 2016-10-19 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | Essential but Not Vulnerable: Indazole Sulfonamides Targeting Inosine Monophosphate Dehydrogenase as Potential Leads against Mycobacterium tuberculosis.
ACS Infect Dis, 3, 2017
|
|
4Z17
 
 | | Thermostable enolase from Chloroflexus aurantiacus | | Descriptor: | Enolase, MAGNESIUM ION, PHOSPHOENOLPYRUVATE | | Authors: | Zadvornyy, O.A, Peters, J.W. | | Deposit date: | 2015-03-26 | | Release date: | 2015-07-01 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Biochemical and Structural Characterization of Enolase from Chloroflexus aurantiacus: Evidence for a Thermophilic Origin.
Front Bioeng Biotechnol, 3, 2015
|
|
5K1L
 
 | |
6VFR
 
 | | Crystal structure of human protocadherin 18 EC1-EC4 | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Harrison, O.J, Brasch, J, Shapiro, L. | | Deposit date: | 2020-01-06 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Family-wide Structural and Biophysical Analysis of Binding Interactions among Non-clustered delta-Protocadherins.
Cell Rep, 30, 2020
|
|
