7ZPB
 
 | | Structure of hemiacetylated human butyrylcholinesterase upon reaction with 8-(3-(4-(prop-2-yn-1-yl)piperazin-1-yl)propoxy)quinoline-2-carbaldehyde | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Denic, M, Chioua, M, Knez, D, Gobec, S, Nachon, F, Marco-Contelles, J.L, Brazzolotto, X. | | Deposit date: | 2022-04-27 | | Release date: | 2023-02-01 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | 8-Hydroxyquinolylnitrones as multifunctional ligands for the therapy of neurodegenerative diseases.
Acta Pharm Sin B, 13, 2023
|
|
1JK4
 
 | | DES 1-6 BOVINE NEUROPHYSIN II COMPLEX WITH VASOPRESSIN | | Descriptor: | CADMIUM ION, Lys Vasopressin, NEUROPHYSIN 2 | | Authors: | Rose, J.P, Wang, B.-C. | | Deposit date: | 2001-07-11 | | Release date: | 2003-02-04 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of an Unliganded Neurophysin and its Vasopressin Complex: Implications for Binding and Allosteric Mechanisms
Protein Sci., 10, 2001
|
|
8P26
 
 | | Crystal structure of Arabidopsis thaliana PAXX | | Descriptor: | U2 small nuclear ribonucleoprotein auxiliary factor-like protein | | Authors: | Ochi, T. | | Deposit date: | 2023-05-15 | | Release date: | 2023-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Plant PAXX has an XLF-like function and stimulates DNA end joining by the Ku-DNA ligase IV/XRCC4 complex.
Plant J., 116, 2023
|
|
7V0J
 
 | | Crystal structure of a CelR catalytic domain active site mutant with bound cellobiose product | | Descriptor: | CALCIUM ION, Glucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Bingman, C.A, Kuch, N, Kutsche, M.E, Parker, A, Smith, R.W, Fox, B.G. | | Deposit date: | 2022-05-10 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Contribution of calcium ligands in substrate binding and product release in the Acetovibrio thermocellus glycoside hydrolase family 9 cellulase CelR.
J.Biol.Chem., 299, 2023
|
|
7V0I
 
 | | Crystal structure of a CelR catalytic domain active site mutant with bound cellohexaose substrate | | Descriptor: | CALCIUM ION, Glucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Bingman, C.A, Kuch, N, Kutsche, M.E, Parker, A, Smith, R.W, Fox, B.G. | | Deposit date: | 2022-05-10 | | Release date: | 2023-04-05 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Contribution of calcium ligands in substrate binding and product release in the Acetovibrio thermocellus glycoside hydrolase family 9 cellulase CelR.
J.Biol.Chem., 299, 2023
|
|
7JT4
 
 | | Crystal Structure of BPTF bromodomain labelled with 5-fluoro-tryptophan | | Descriptor: | Nucleosome-remodeling factor subunit BPTF | | Authors: | Johnson, J.A, Shi, K, Aihara, H, Pomerantz, W.C.K. | | Deposit date: | 2020-08-17 | | Release date: | 2021-07-28 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Selectivity, ligand deconstruction, and cellular activity analysis of a BPTF bromodomain inhibitor
Org.Biomol.Chem., 17, 2019
|
|
1USK
 
 | | L-leucine-binding protein with leucine bound | | Descriptor: | LEUCINE, LEUCINE-SPECIFIC BINDING PROTEIN | | Authors: | Magnusson, U, Salopek-Sondi, B, Luck, L.A, Mowbray, S.L. | | Deposit date: | 2003-11-25 | | Release date: | 2003-12-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | X-Ray Structures of the Leucine-Binding Protein Illustrate Conformational Changes and the Basis of Ligand Specificity
J.Biol.Chem., 279, 2004
|
|
1USG
 
 | | L-leucine-binding protein, apo form | | Descriptor: | LEUCINE-SPECIFIC BINDING PROTEIN | | Authors: | Magnusson, U, Salopek-Sondi, B, Luck, L.A, Mowbray, S.L. | | Deposit date: | 2003-11-24 | | Release date: | 2003-12-18 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | X-Ray Structures of the Leucine-Binding Protein Illustrate Conformational Changes and the Basis of Ligand Specificity
J.Biol.Chem., 279, 2004
|
|
2G5V
 
 | |
4GIZ
 
 | | Crystal structure of full-length human papillomavirus oncoprotein E6 in complex with LXXLL peptide of ubiquitin ligase E6AP at 2.55 A resolution | | Descriptor: | Maltose-binding periplasmic protein, UBIQUITIN LIGASE EA6P: chimeric protein, Protein E6, ... | | Authors: | McEwen, A.G, Zanier, K, Charbonnier, S, Poussin, P, Cura, V, Vande Pol, S, Trave, G, Cavarelli, J. | | Deposit date: | 2012-08-09 | | Release date: | 2013-01-23 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural basis for hijacking of cellular LxxLL motifs by papillomavirus E6 oncoproteins.
Science, 339, 2013
|
|
1J06
 
 | | Crystal structure of mouse acetylcholinesterase in the apo form | | Descriptor: | 2-(2-ETHOXYETHOXY)ETHANOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, CARBONATE ION, ... | | Authors: | Bourne, Y, Taylor, P, Radic, Z, Marchot, P. | | Deposit date: | 2002-11-07 | | Release date: | 2003-02-04 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural insights into ligand interactions at the acetylcholinesterase peripheral anionic site
EMBO J., 22, 2003
|
|
7V12
 
 | | Factor XIa in Complex with Compound 2f | | Descriptor: | 5-[1-[(1~{R})-1-[5-[3-chloranyl-2-fluoranyl-6-(1,2,3,4-tetrazol-1-yl)phenyl]-1-oxidanyl-pyridin-2-yl]-2-cyclopropyl-ethyl]pyrazol-4-yl]-6-fluoranyl-pyridin-2-amine, CITRIC ACID, Coagulation factor XIa light chain | | Authors: | Shaffer, P.L, Spurlino, J, Milligan, C.M. | | Deposit date: | 2022-05-11 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.631 Å) | | Cite: | Discovery of Potent and Orally Bioavailable Pyridine N-Oxide-Based Factor XIa Inhibitors through Exploiting Nonclassical Interactions.
J.Med.Chem., 65, 2022
|
|
7V10
 
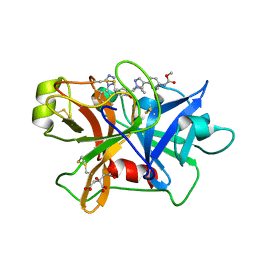 | | Factor XIa in Complex with Compound 2d | | Descriptor: | CITRIC ACID, Coagulation factor XIa light chain, methyl ~{N}-[4-[1-[(1~{R})-1-[5-[3-chloranyl-2-fluoranyl-6-(1,2,3,4-tetrazol-1-yl)phenyl]-1-oxidanyl-pyridin-2-yl]-2-cyclopropyl-ethyl]pyrazol-4-yl]phenyl]carbamate | | Authors: | Shaffer, P.L, Spurlino, J, Milligan, C.M. | | Deposit date: | 2022-05-11 | | Release date: | 2022-08-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Discovery of Potent and Orally Bioavailable Pyridine N-Oxide-Based Factor XIa Inhibitors through Exploiting Nonclassical Interactions.
J.Med.Chem., 65, 2022
|
|
1J4R
 
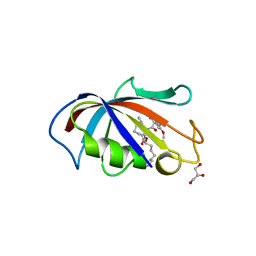 | | FK506 BINDING PROTEIN COMPLEXED WITH FKB-001 | | Descriptor: | 1-[2,2-DIFLUORO-2-(3,4,5-TRIMETHOXY-PHENYL)-ACETYL]-PIPERIDINE-2-CARBOXYLIC ACID 4-PHENYL-1-(3-PYRIDIN-3-YL-PROPYL)-BUTYL ESTER, FK506-BINDING PROTEIN, GLYCEROL, ... | | Authors: | Sheriff, S. | | Deposit date: | 2001-10-29 | | Release date: | 2001-12-19 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | 2-Aryl-2,2-difluoroacetamide FKBP12 ligands: synthesis and X-ray structural studies.
Org.Lett., 3, 2001
|
|
6K1Z
 
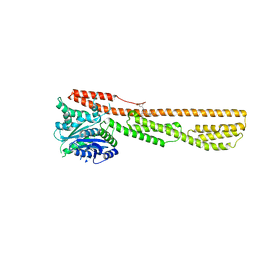 | | Crystal structure of farnesylated hGBP1 | | Descriptor: | FARNESYL, Guanylate-binding protein 1 | | Authors: | Du, S, Xiao, J.Y. | | Deposit date: | 2019-05-13 | | Release date: | 2019-06-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.307 Å) | | Cite: | Structural mechanism for guanylate-binding proteins (GBPs) targeting by the Shigella E3 ligase IpaH9.8.
Plos Pathog., 15, 2019
|
|
1W6U
 
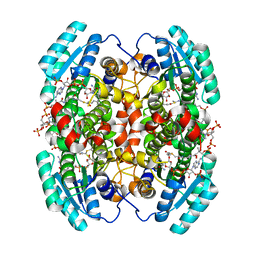 | | Structure of human DECR ternary complex | | Descriptor: | 2,4-DIENOYL-COA REDUCTASE, MITOCHONDRIAL PRECURSOR, HEXANOYL-COENZYME A, ... | | Authors: | Alphey, M.S, Byres, E, Li, D, Hunter, W.N. | | Deposit date: | 2004-08-24 | | Release date: | 2004-10-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and Reactivity of Human Mitochondrial 2,4-Dienoyl-Coa Reductase: Enzyme-Ligand Interactions in a Distinctive Short-Chain Reductase Active Site
J.Biol.Chem., 280, 2005
|
|
1W73
 
 | | Binary structure of human DECR solved by SeMet SAD. | | Descriptor: | 2,4-DIENOYL-COA REDUCTASE, GLYCEROL, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Alphey, M.S, Byres, E, Hunter, W.N. | | Deposit date: | 2004-08-27 | | Release date: | 2004-10-28 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure and Reactivity of Human Mitochondrial 2,4-Dienoyl-Coa Reductase: Enzyme-Ligand Interactions in a Distinctive Short-Chain Reductase Active Site
J.Biol.Chem., 280, 2005
|
|
2H28
 
 | | Crystal structure of YeeU from E. coli. Northeast Structural Genomics target ER304 | | Descriptor: | CHLORIDE ION, GLYCEROL, Hypothetical protein yeeU, ... | | Authors: | Arbing, M, Su, M, Benach, J, Karpowich, N.K, Jiang, M, Xiao, R, Cunningham, K, Ma, L.-C, Chen, C.X, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-05-18 | | Release date: | 2006-07-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structures of Phd-Doc, HigA, and YeeU Establish Multiple Evolutionary Links between Microbial Growth-Regulating Toxin-Antitoxin Systems.
Structure, 18, 2010
|
|
1J07
 
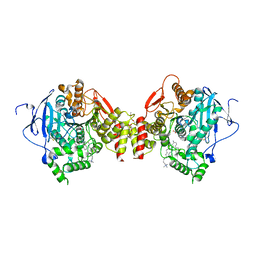 | | Crystal structure of the mouse acetylcholinesterase-decidium complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 3,8-DIAMINO-5,10'-(TRIMETHYLAMMONIUM)DECYL-6-PHENYL PHENANTHRIDINIUM, CARBONATE ION, ... | | Authors: | Bourne, Y, Taylor, P, Radic, Z, Marchot, P. | | Deposit date: | 2002-11-07 | | Release date: | 2003-02-04 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural insights into ligand interactions at the acetylcholinesterase peripheral anionic site
EMBO J., 22, 2003
|
|
2ZEX
 
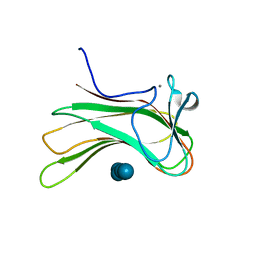 | | Family 16 carbohydrate binding module | | Descriptor: | CALCIUM ION, S-layer associated multidomain endoglucanase, beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose-(1-4)-beta-D-glucopyranose | | Authors: | Bae, B, Nair, S.K. | | Deposit date: | 2007-12-18 | | Release date: | 2008-03-04 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Molecular Basis for the Selectivity and Specificity of Ligand Recognition by the Family 16 Carbohydrate-binding Modules from Thermoanaerobacterium polysaccharolyticum ManA
J.Biol.Chem., 283, 2008
|
|
3D10
 
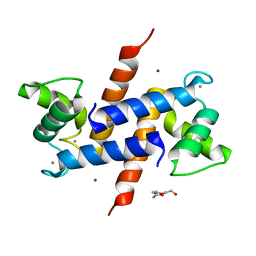 | | Crystal Structure of S100B in the Calcium and Zinc Loaded State at pH 10.0 | | Descriptor: | CALCIUM ION, Protein S100-B, TRIETHYLENE GLYCOL, ... | | Authors: | Ostendorp, T, Diez, J, Heizmann, C.W, Fritz, G. | | Deposit date: | 2008-05-02 | | Release date: | 2009-04-14 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | The crystal structures of human S100B in the zinc- and calcium-loaded state at three pH values reveal zinc ligand swapping.
Biochim.Biophys.Acta, 1813, 2011
|
|
2CDT
 
 | | alpha-SPECTRIN SH3 DOMAIN A56S MUTANT | | Descriptor: | SPECTRIN ALPHA CHAIN | | Authors: | Casares, S, Camara-Artigas, A, Vega, M.C, Lopez-Mayorga, O, Conejero-Lara, F. | | Deposit date: | 2006-01-27 | | Release date: | 2007-02-20 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.54 Å) | | Cite: | Cooperative Propagation of Local Stability Changes from Low-Stability and High-Stability Regions in a SH3 Domain.
Proteins: Struct., Funct., Bioinf., 67, 2007
|
|
1LR8
 
 | |
8CNP
 
 | |
4I7J
 
 | | T4 Lysozyme L99A/M102H with benzene bound | | Descriptor: | 2-HYDROXYETHYL DISULFIDE, ACETATE ION, BENZENE, ... | | Authors: | Merski, M, Shoichet, B.K. | | Deposit date: | 2012-11-30 | | Release date: | 2013-03-27 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | The impact of introducing a histidine into an apolar cavity site on docking and ligand recognition.
J.Med.Chem., 56, 2013
|
|
