8VAQ
 
 | |
8C6W
 
 | | PBP AccA from A. tumefaciens Bo542 in apoform 1 | | Descriptor: | Agrocinopine utilization periplasmic binding protein AccA | | Authors: | Morera, S, Vigouroux, A, Legrand, P. | | Deposit date: | 2023-01-12 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
7AYH
 
 | | Crystal structure of Aurora A in complex with 7-(2-Anilinopyrimidin-4-yl)-1-benzazepin-2-one derivative (compound 2c) | | Descriptor: | 7-[2-[(4-methoxyphenyl)amino]pyrimidin-4-yl]-1,3,4,5-tetrahydro-1-benzazepin-2-one, Aurora kinase A | | Authors: | Chaikuad, A, Karatas, M, Kunick, C, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2020-11-12 | | Release date: | 2021-01-13 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | 7-(2-Anilinopyrimidin-4-yl)-1-benzazepin-2-ones Designed by a "Cut and Glue" Strategy Are Dual Aurora A/VEGF-R Kinase Inhibitors.
Molecules, 26, 2021
|
|
6HRG
 
 | | Structure of Igni18, a novel metallo hydrolase from the hyperthermophilic archaeon Ignicoccus hospitalis KIN4/I | | Descriptor: | PHOSPHATE ION, POTASSIUM ION, UPF0173 metal-dependent hydrolase Igni_1254, ... | | Authors: | Smits, S.H, Streit, W.R, Jaeger, K.E, Hoeppner, A. | | Deposit date: | 2018-09-26 | | Release date: | 2019-10-09 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | A promiscuous ancestral enzyme ́s structure unveils protein variable regions of the highly diverse metallo-beta-lactamase family.
Commun Biol, 4, 2021
|
|
6Z8C
 
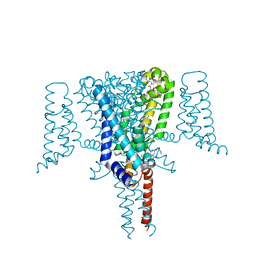 | | Crystal Structure of the Voltage-Gated Sodium Channel NavMs (F208L) in complex with N-desmethyltamoxifen (3.2 A resolution) | | Descriptor: | 2-[4-[(~{Z})-1,2-diphenylbut-1-enyl]phenoxy]-~{N}-methyl-ethanamine, DODECAETHYLENE GLYCOL, HEGA-10, ... | | Authors: | Sula, A, Hollingworth, D, Wallace, B.A. | | Deposit date: | 2020-06-02 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | A tamoxifen receptor within a voltage-gated sodium channel.
Mol.Cell, 81, 2021
|
|
8CNB
 
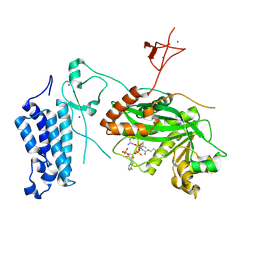 | | Crystal structure of CREBBP-Y1503C histone acetyltransferase domain in complex with Coenzyme A | | Descriptor: | COENZYME A, ZINC ION, histone acetyltransferase | | Authors: | Mechaly, A.E, Zhang, W, Haouz, A, Green, M, Rodrigues-Lima, F. | | Deposit date: | 2023-02-22 | | Release date: | 2024-03-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.986 Å) | | Cite: | Crystal structure of CREBBP-Y1503C histone acetyltransferase domain in complex with Coenzyme A
To Be Published
|
|
8CMZ
 
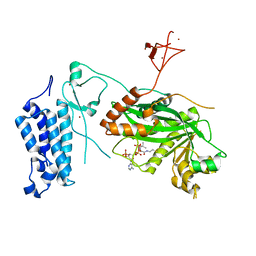 | | Crystal structure of CREBBP-R1446C histone acetyltransferase domain in complex with Coenzyme A | | Descriptor: | COENZYME A, ZINC ION, histone acetyltransferase | | Authors: | Mechaly, A.E, Zhang, W, Haouz, A, Green, M, Rodrigues-Lima, F. | | Deposit date: | 2023-02-21 | | Release date: | 2024-03-06 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.252 Å) | | Cite: | Crystal structure of CREBBP-R1446C histone acetyltransferase domain in complex with Coenzyme A
To Be Published
|
|
4YYM
 
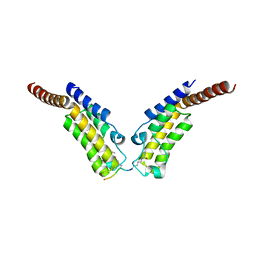 | | Crystal structure of TAF1 BD2 Bromodomain bound to a butyryllysine peptide | | Descriptor: | CALCIUM ION, Histone H4, Transcription initiation factor TFIID subunit 1 | | Authors: | Tang, Y, Bellon, S, Cochran, A.G, Poy, F. | | Deposit date: | 2015-03-24 | | Release date: | 2015-09-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | A Subset of Human Bromodomains Recognizes Butyryllysine and Crotonyllysine Histone Peptide Modifications.
Structure, 23, 2015
|
|
6ONE
 
 | | Crystal structure of HIV-1 LM/HT Clade A/E CRF01 gp120 core in complex with (S)-MCG-III-188-A01. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, clade A/E 93TH057 HIV-1 gp120 core, ... | | Authors: | Tolbert, W.D, Sherburn, R, Pazgier, M. | | Deposit date: | 2019-04-22 | | Release date: | 2019-10-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A New Family of Small-Molecule CD4-Mimetic Compounds Contacts Highly Conserved Aspartic Acid 368 of HIV-1 gp120 and Mediates Antibody-Dependent Cellular Cytotoxicity.
J.Virol., 93, 2019
|
|
6ONH
 
 | | Crystal structure of HIV-1 LM/HT Clade A/E CRF01 gp120 core in complex with (S)-MCG-IV-031-A05. | | Descriptor: | (3S)-N~3~-(4-chloro-3-fluorophenyl)-N~1~-propylpiperidine-1,3-dicarboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Tolbert, W.D, Sherburn, R, Pazgier, M. | | Deposit date: | 2019-04-22 | | Release date: | 2019-10-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.244 Å) | | Cite: | A New Family of Small-Molecule CD4-Mimetic Compounds Contacts Highly Conserved Aspartic Acid 368 of HIV-1 gp120 and Mediates Antibody-Dependent Cellular Cytotoxicity.
J.Virol., 93, 2019
|
|
6ONV
 
 | | Crystal structure of HIV-1 LM/HT Clade A/E CRF01 gp120 core in complex with (S)-MCG-III-027-D05. | | Descriptor: | (3S)-N-(4-chloro-3-fluorophenyl)-1-(methylsulfonyl)piperidine-3-carboxamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ... | | Authors: | Tolbert, W.D, Sherburn, R, Pazgier, M. | | Deposit date: | 2019-04-22 | | Release date: | 2019-10-23 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.253 Å) | | Cite: | A New Family of Small-Molecule CD4-Mimetic Compounds Contacts Highly Conserved Aspartic Acid 368 of HIV-1 gp120 and Mediates Antibody-Dependent Cellular Cytotoxicity.
J.Virol., 93, 2019
|
|
8CH2
 
 | | PBP AccA from A. vitis S4 in complex with L-arabinose-2-phosphate (A2P) | | Descriptor: | 1,2-ETHANEDIOL, 2-O-phosphono-alpha-L-arabinopyranose, 2-O-phosphono-beta-L-arabinopyranose, ... | | Authors: | Morera, S, Deicsics, G, Vigouroux, A. | | Deposit date: | 2023-02-06 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.404 Å) | | Cite: | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
8CJU
 
 | | PBP AccA from A. vitis S4 in complex with agrocin84 | | Descriptor: | ABC transporter substrate binding protein (Agrocinopine), [(2S,3R,4S,5S,6R)-6-(hydroxymethyl)-2,4,5-tris(oxidanyl)oxan-3-yl]oxy-N-[9-[(2R,3S,5R)-5-[[[(2R,3S)-4-methyl-2,3-bis(oxidanyl)pentanoyl]amino]-oxidanyl-phosphoryl]oxy-3-oxidanyl-oxolan-2-yl]purin-6-yl]phosphonamidic acid | | Authors: | Morera, S, Vigouroux, A. | | Deposit date: | 2023-02-13 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.349 Å) | | Cite: | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
8CHC
 
 | | PBP AccA from A. vitis S4 in complex with Agrocinopine D-like | | Descriptor: | Agrocinopine D-like (C2-C2 linked; with two alpha-D-glucopyranoses), Agrocinopine utilization periplasmic binding protein AccA | | Authors: | Morera, S, Vigouroux, A, Deicsics, G. | | Deposit date: | 2023-02-07 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.679 Å) | | Cite: | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
8CI6
 
 | | PBP AccA from A. vitis S4 in complex with D-glucose-2-phosphate (G2P) | | Descriptor: | 2-O-phosphono-alpha-D-glucopyranose, 2-O-phosphono-beta-D-glucopyranose, ABC transporter substrate binding protein (Agrocinopine) | | Authors: | Morera, S, Vigouroux, A. | | Deposit date: | 2023-02-08 | | Release date: | 2024-01-24 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.199 Å) | | Cite: | A highly conserved ligand-binding site for AccA transporters of antibiotic and quorum-sensing regulator in Agrobacterium leads to a different specificity.
Biochem.J., 481, 2024
|
|
3N6S
 
 | | Crystal structure of human mitochondrial mTERF in complex with a 15-mer DNA encompassing the tRNALeu(UUR) binding sequence | | Descriptor: | DNA (5'-D(*AP*TP*GP*GP*CP*AP*GP*AP*GP*CP*CP*CP*GP*GP*T)-3'), DNA (5'-D(*TP*AP*CP*CP*GP*GP*GP*CP*TP*CP*TP*GP*CP*CP*A)-3'), Transcription termination factor, ... | | Authors: | Jimenez-Menendez, N, Fernandez-Millan, P, Rubio-Cosials, A, Arnan, C, Montoya, J, Jacobs, H.T, Bernado, P, Coll, M, Uson, I, Sola, M. | | Deposit date: | 2010-05-26 | | Release date: | 2010-06-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Human mitochondrial mTERF wraps around DNA through a left-handed superhelical tandem repeat.
Nat.Struct.Mol.Biol., 17, 2010
|
|
8V4R
 
 | |
7KU0
 
 | | Data clustering and dynamics of chymotrypsinogen cluster 138 (yellow) structure | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-10 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7KU2
 
 | | Data clustering and dynamics of chymotrypsinogen clulster 140 (structure) | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.185 Å) | | Cite: | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7KU3
 
 | | Data clustering and dynamics of chymotrypsinogen cluster 141 (cyan) structure | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-10 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7KTZ
 
 | | Data clustering and dynamics of chymotrypsinogen cluster 131 (purple) structure | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-10 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
7KTY
 
 | | Data clustering and dynamics of chymotrypsinogen average structure | | Descriptor: | Chymotrypsinogen A, SULFATE ION | | Authors: | Nguyen, T, Phan, K.L, Kreitler, D.F, Andrews, L.C, Gabelli, S.B, Kozakov, D, Jakoncic, J, Shi, W, Sweet, R.M, Soares, A.S, Bernstein, H.J. | | Deposit date: | 2020-11-24 | | Release date: | 2021-03-10 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A simple technique to classify diffraction data from dynamic proteins according to individual polymorphs.
Acta Crystallogr D Struct Biol, 78, 2022
|
|
6RQK
 
 | | Crystal structure of GH125 1,6-alpha-mannosidase from Clostridium perfringens in complex with mannoimidazole | | Descriptor: | (5R,6R,7S,8R)-5-(HYDROXYMETHYL)-5,6,7,8-TETRAHYDROIMIDAZO[1,2-A]PYRIDINE-6,7,8-TRIOL, Alpha-1,6-mannosidase | | Authors: | Males, A, Davies, G.J. | | Deposit date: | 2019-05-16 | | Release date: | 2019-08-28 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Distortion of mannoimidazole supports a B2,5boat transition state for the family GH125 alpha-1,6-mannosidase from Clostridium perfringens.
Org.Biomol.Chem., 17, 2019
|
|
6PMB
 
 | | TRK-A IN COMPLEX WITH LIGAND 1a | | Descriptor: | 2-[5,7-dimethyl-2-(pyridin-3-yl)[1,2,4]triazolo[1,5-a]pyrimidin-6-yl]-N-[3-(trifluoromethyl)phenyl]acetamide, High affinity nerve growth factor receptor | | Authors: | Subramanian, G, Brown, D.G. | | Deposit date: | 2019-07-01 | | Release date: | 2020-02-26 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Synthetic inhibitor leads of human tropomyosin receptor kinase A ( h TrkA).
Rsc Med Chem, 11, 2020
|
|
8UT7
 
 | | CryoEM structure of A/Perth/16/2009 H3 in complex with polyclonal Fab from mice immunized with H3 stem nanoparticles-28 days post immunization | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, H3D28 pFab HC Fv_polyA, ... | | Authors: | Huang, J, Han, J, Ward, A.B. | | Deposit date: | 2023-10-30 | | Release date: | 2024-05-08 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Eliciting a single amino acid change by vaccination generates antibody protection against group 1 and group 2 influenza A viruses.
Immunity, 57, 2024
|
|
