2OW6
 
 | | Golgi alpha-mannosidase II complex with (1r,5s,6s,7r,8s)-1-thioniabicyclo[4.3.0]nonan-5,7,8-triol chloride | | Descriptor: | (1R,5S,6S,7R,8S)-1-THIONIABICYCLO[4.3.0]NONAN-5,7,8-TRIOL, (4R)-2-METHYLPENTANE-2,4-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kuntz, D.A. | | Deposit date: | 2007-02-15 | | Release date: | 2008-01-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.19 Å) | | Cite: | Binding of sulfonium-ion analogues of di-epi-swainsonine and 8-epi-lentiginosine to Drosophila Golgi alpha-mannosidase II: The role of water in inhibitor binding.
Proteins, 71, 2008
|
|
2DND
 
 | | A BIFURCATED HYDROGEN-BONDED CONFORMATION IN THE D(A.T) BASE PAIRS OF THE DNA DODECAMER D(CGCAAATTTGCG) AND ITS COMPLEX WITH DISTAMYCIN | | Descriptor: | DISTAMYCIN A, DNA (5'-D(*CP*GP*CP*AP*AP*AP*TP*TP*TP*GP*CP*G)-3') | | Authors: | Coll, M, Frederick, C.A, Wang, A.H.-J, Rich, A. | | Deposit date: | 1988-08-29 | | Release date: | 1989-01-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | A bifurcated hydrogen-bonded conformation in the d(A.T) base pairs of the DNA dodecamer d(CGCAAATTTGCG) and its complex with distamycin.
Proc.Natl.Acad.Sci.USA, 84, 1987
|
|
2OW7
 
 | | Golgi alpha-mannosidase II complex with (1R,6S,7R,8S)-1-thioniabicyclo[4.3.0]nonan-7,8-diol chloride | | Descriptor: | (1R,6S,7R,8S)-1-THIONIABICYCLO[4.3.0]NONAN-7,8-DIOL, (4R)-2-METHYLPENTANE-2,4-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Kuntz, D.A. | | Deposit date: | 2007-02-15 | | Release date: | 2008-01-08 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Binding of sulfonium-ion analogues of di-epi-swainsonine and 8-epi-lentiginosine to Drosophila Golgi alpha-mannosidase II: The role of water in inhibitor binding.
Proteins, 71, 2008
|
|
1CAU
 
 | | DETERMINATION OF THREE CRYSTAL STRUCTURES OF CANAVALIN BY MOLECULAR REPLACEMENT | | Descriptor: | CANAVALIN | | Authors: | Ko, T-P, Ng, J.D, Day, J, Greenwood, A, McPherson, A. | | Deposit date: | 1993-07-08 | | Release date: | 1993-10-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Determination of three crystal structures of canavalin by molecular replacement.
Acta Crystallogr.,Sect.D, 49, 1993
|
|
1CAV
 
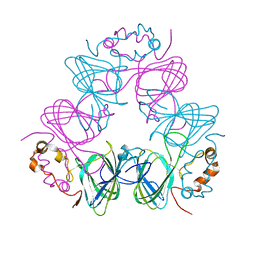 | |
1C9Z
 
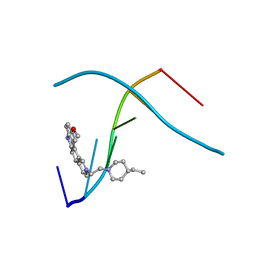 | | D232-CGTACG | | Descriptor: | 1,3-DI[[[10-METHOXY-7H-PYRIDO[4,3-C]CARBAZOL-2-IUMYL]-ETHYL]-PIPERIDIN-4-YL]-PROPANE, 5'-D(*CP*GP*TP*AP*CP*G)-3' | | Authors: | Williams, L.D. | | Deposit date: | 1999-08-03 | | Release date: | 2000-04-02 | | Last modified: | 2024-04-17 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Effects of cationic charge on three-dimensional structures of intercalative complexes: structure of a bis-intercalated DNA complex solved by MAD phasing.
Curr.Med.Chem., 7, 2000
|
|
2LMS
 
 | | A single GalNAc residue on Threonine-106 modifies the dynamics and the structure of Interferon alpha-2a around the glycosylation site | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, Interferon alpha-2 | | Authors: | Ghasriani, H, Belcourt, P.J.F, Sauve, S, Hodgson, D.J, Gingras, G, Brochu, D, Gilbert, M, Aubin, Y. | | Deposit date: | 2011-12-12 | | Release date: | 2012-12-05 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A single N-acetylgalactosamine residue at threonine 106 modifies the dynamics and structure of interferon alpha2a around the glycosylation site.
J.Biol.Chem., 288, 2013
|
|
2LI2
 
 | | Mono-O-GalNAc glycosylated Mucin sequence based on MUC2 Mucin glycoprotein tandem repeat | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose, MUC2 Mucin Domain Peptide | | Authors: | Borgert, A, Heimburg-Molinaro, J, Lasanajak, Y, Ju, T, Liu, M, Thompson, P, Ragupathi, G, Barany, G, Cummings, R, Smith, D, Live, D. | | Deposit date: | 2011-08-18 | | Release date: | 2012-04-04 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Deciphering structural elements of mucin glycoprotein recognition.
Acs Chem.Biol., 7, 2012
|
|
1QKD
 
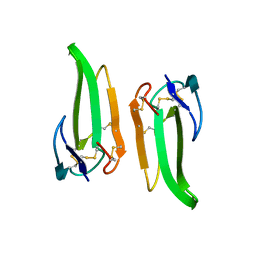 | | ERABUTOXIN | | Descriptor: | ERABUTOXIN A | | Authors: | Nastopoulos, V, Kanellopoulos, P.N, Tsernoglou, D. | | Deposit date: | 1998-01-16 | | Release date: | 1999-02-16 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | Structure of dimeric and monomeric erabutoxin a refined at 1.5 A resolution.
Acta Crystallogr.,Sect.D, 54, 1998
|
|
1QSX
 
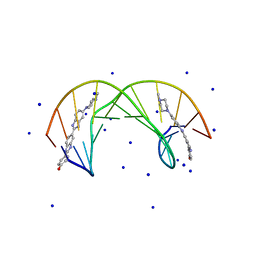 | | SOLUTION NMR STRUCTURE OF THE 2:1 HOECHST 33258-D(CTTTTGCAAAAG)2 COMPLEX | | Descriptor: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, 5'-D(CP*TP*TP*TP*TP*GP*CP*AP*AP*AP*AP*G)-3', SODIUM ION | | Authors: | Gavathiotis, E, Sharman, G.J, Searle, M.S. | | Deposit date: | 1999-06-24 | | Release date: | 2000-02-07 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Sequence-dependent variation in DNA minor groove width dictates orientational preference of Hoechst 33258 in A-tract recognition: solution NMR structure of the 2:1 complex with d(CTTTTGCAAAAG)(2).
Nucleic Acids Res., 28, 2000
|
|
1QCH
 
 | |
2BWB
 
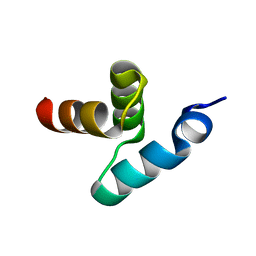 | | Crystal structure of the UBA domain of Dsk2 from S. cerevisiae | | Descriptor: | UBIQUITIN-LIKE PROTEIN DSK2 | | Authors: | Lowe, E.D, Hasan, N, Trempe, J.-F, Fonso, L, Noble, M.E.M, Endicott, J.A, Johnson, L.N, Brown, N.R. | | Deposit date: | 2005-07-13 | | Release date: | 2006-01-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of the Dsk2 Ubl and Uba Domains and Their Complex.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
3OIA
 
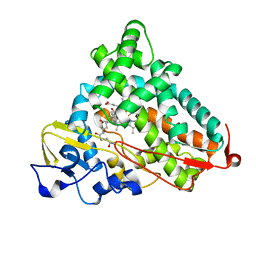 | | Crystal Structure of Cytochrome P450cam crystallized in the presence of a tethered substrate analog AdaC1-C8GluEtg-Bio | | Descriptor: | Camphor 5-monooxygenase, N-{15-oxo-19-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]-4,7,10-trioxa-14-azanonadec-1-yl}-N'-(8-{[(3S,5S,7S)-tricyclo[3.3.1.1~3,7~]dec-1-ylcarbonyl]amino}octyl)pentanediamide, POTASSIUM ION, ... | | Authors: | Lee, Y.-T, Wilson, R.F, Glazer, E.C, Goodin, D.B. | | Deposit date: | 2010-08-18 | | Release date: | 2010-11-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Crystal Structure of Cytochrome P450cam crystallized in the presence of a tethered substrate analog AdaC1-C8GluEtg-Bio
To be Published
|
|
2G1Z
 
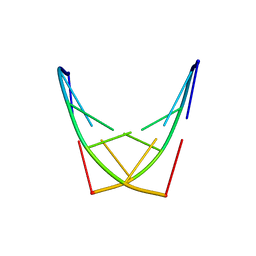 | |
1QC1
 
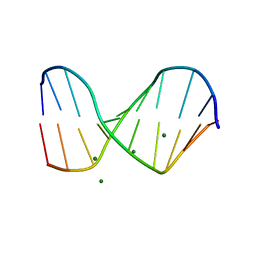 | |
1DN8
 
 | |
1DNH
 
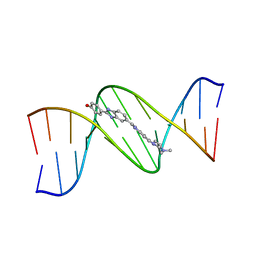 | | THE MOLECULAR STRUCTURE OF THE COMPLEX OF HOECHST 33258 AND THE DNA DODECAMER D(CGCGAATTCGCG) | | Descriptor: | 2'-(4-HYDROXYPHENYL)-5-(4-METHYL-1-PIPERAZINYL)-2,5'-BI-BENZIMIDAZOLE, DNA (5'-D(*CP*GP*CP*GP*AP*AP*TP*TP*CP*GP*CP*G)-3') | | Authors: | Teng, M.-K, Usman, N, Frederick, C.A, Wang, A.H.-J. | | Deposit date: | 1988-02-16 | | Release date: | 1989-01-09 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | The molecular structure of the complex of Hoechst 33258 and the DNA dodecamer d(CGCGAATTCGCG).
Nucleic Acids Res., 16, 1988
|
|
3PJ2
 
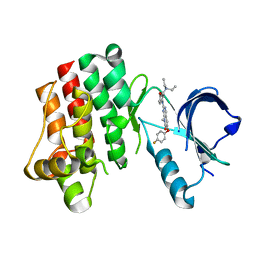 | |
1Q2X
 
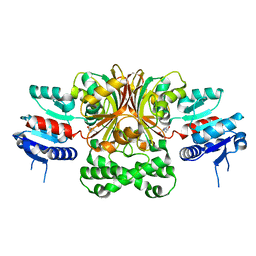 | | Crystal Structure of the E243D Mutant of Aspartate Semialdehyde Dehydrogenase from Haemophilus influenzae bound with substrate aspartate semialdehyde | | Descriptor: | Aspartate-semialdehyde dehydrogenase | | Authors: | Blanco, J, Moore, R.A, Faehnle, C.R, Coe, D.M, Viola, R.E. | | Deposit date: | 2003-07-26 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The role of substrate-binding groups in the mechanism of aspartate-beta-semialdehyde dehydrogenase.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
1PL1
 
 | | Crystal structure of human glutathione transferase (GST) A1-1 in complex with a decarboxy-glutathione | | Descriptor: | CHLORIDE ION, Glutathione S-transferase A1, N-(4-AMINOBUTANOYL)-S-(4-METHOXYBENZYL)-L-CYSTEINYLGLYCINE | | Authors: | Grahn, E, Jakobsson, E, Gustafsson, A, Grehn, L, Olin, B, Wahlberg, M, Madsen, D, Kleywegt, G.J, Mannervik, B. | | Deposit date: | 2003-06-06 | | Release date: | 2004-06-22 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | New crystal structures of human glutathione transferase A1-1 shed light on glutathione binding and the conformation of the C-terminal helix.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
1PXD
 
 | | Crystal structure of the complex of jacalin with meso-tetrasulphonatophenylporphyrin. | | Descriptor: | 5,10,15,20-TETRAKIS(4-SULPFONATOPHENYL)-21H,23H-PORPHINE, Agglutinin alpha chain, Agglutinin beta-3 chain | | Authors: | Goel, M, Anuradha, P, Kaur, K.J, Maiya, B.G, Swamy, M.J, Salunke, D.M. | | Deposit date: | 2003-07-03 | | Release date: | 2004-02-03 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Porphyrin binding to jacalin is facilitated by the inherent plasticity of the carbohydrate-binding site: novel mode of lectin-ligand interaction.
Acta Crystallogr.,Sect.D, 60, 2004
|
|
2HTA
 
 | | Crystal Structure of a putative mutarotase (YeaD) from Salmonella typhimurium in orthorhombic form | | Descriptor: | GLYCEROL, Putative enzyme related to aldose 1-epimerase, SULFATE ION | | Authors: | Chittori, S, Simanshu, D.K, Savithri, H.S, Murthy, M.R.N. | | Deposit date: | 2006-07-25 | | Release date: | 2007-01-23 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the putative mutarotase YeaD from Salmonella typhimurium: structural comparison with galactose mutarotases.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
2XBD
 
 | | INTERNAL XYLAN BINDING DOMAIN FROM CELLULOMONAS FIMI XYLANASE D, NMR, MINIMIZED AVERAGE STRUCTURE | | Descriptor: | XYLANASE D | | Authors: | Simpson, P.J, Bolam, D.N, Cooper, A, Ciruela, A, Hazlewood, G.P, Gilbert, H.J, Williamson, M.P. | | Deposit date: | 1998-10-27 | | Release date: | 1999-07-21 | | Last modified: | 2022-03-16 | | Method: | SOLUTION NMR | | Cite: | A family IIb xylan-binding domain has a similar secondary structure to a homologous family IIa cellulose-binding domain but different ligand specificity.
Structure Fold.Des., 7, 1999
|
|
2JFG
 
 | | Crystal structure of MurD ligase in complex with UMA and ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, SULFATE ION, UDP-N-ACETYLMURAMOYLALANINE--D-GLUTAMATE LIGASE, ... | | Authors: | Kotnik, M, Humljan, J, Contreras-Martel, C, Oblak, M, Kristan, K, Herve, M, Blanot, D, Urleb, U, Gobec, S, Dessen, A, Solmajer, T. | | Deposit date: | 2007-02-01 | | Release date: | 2007-05-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.52 Å) | | Cite: | Structural and functional characterization of enantiomeric glutamic acid derivatives as potential transition state analogue inhibitors of MurD ligase.
J. Mol. Biol., 370, 2007
|
|
2JFH
 
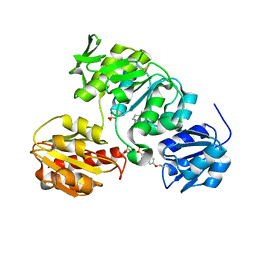 | | Crystal structure of MurD ligase in complex with L-Glu containing sulfonamide inhibitor | | Descriptor: | N-[(6-BUTOXYNAPHTHALEN-2-YL)SULFONYL]-L-GLUTAMIC ACID, SULFATE ION, UDP-N-ACETYLMURAMOYL-ALANINE-D-GLUTAMATE LIGASE | | Authors: | Kotnik, M, Humljan, J, Contreras-Martel, C, Oblak, M, Kristan, K, Herve, M, Blanot, D, Urleb, U, Gobec, S, Dessen, A, Solmajer, T. | | Deposit date: | 2007-02-01 | | Release date: | 2007-05-15 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Structural and Functional Characterization of Enantiomeric Glutamic Acid Derivatives as Potential Transition State Analogue Inhibitors of Murd Ligase.
J.Mol.Biol., 370, 2007
|
|
