4UXQ
 
 | | FGFR4 in complex with Ponatinib | | Descriptor: | 3-(imidazo[1,2-b]pyridazin-3-ylethynyl)-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzam ide, FIBROBLAST GROWTH FACTOR RECEPTOR 4, SULFATE ION | | Authors: | Tucker, J, Klein, T, Breed, J, Breeze, A, Overman, R, Phillips, C, Norman, R.A. | | Deposit date: | 2014-08-27 | | Release date: | 2014-12-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural Insights Into Fgfr Kinase Isoform Selectivity: Diverse Binding Modes of Azd4547 and Ponatinib in Complex with Fgfr1 and Fgfr4
Structure, 22, 2014
|
|
4UU6
 
 | | CRYSTAL STRUCTURE OF THE PDZ DOMAIN OF PALS1 | | Descriptor: | ACETATE ION, CHLORIDE ION, GLYCEROL, ... | | Authors: | Ivanova, M.E, Purkiss, A.G, McDonald, N.Q. | | Deposit date: | 2014-07-24 | | Release date: | 2015-01-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structures of the human Pals1 PDZ domain with and without ligand suggest gated access of Crb to the PDZ peptide-binding groove.
Acta Crystallogr. D Biol. Crystallogr., 71, 2015
|
|
4UVK
 
 | |
7YPL
 
 | |
7YRV
 
 | |
4UZR
 
 | |
6ES8
 
 | | HIV capsid hexamer with IP6 ligand | | Descriptor: | Gag protein, INOSITOL HEXAKISPHOSPHATE | | Authors: | James, L.C. | | Deposit date: | 2017-10-19 | | Release date: | 2018-08-15 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | IP6 is an HIV pocket factor that prevents capsid collapse and promotes DNA synthesis.
Elife, 7, 2018
|
|
4UR0
 
 | | Crystal structure of the PCE reductive dehalogenase from S. multivorans in complex with trichloroethene | | Descriptor: | 1,1,2-trichloroethene, BENZAMIDINE, GLYCEROL, ... | | Authors: | Bommer, M, Kunze, C, Fesseler, J, Schubert, T, Diekert, G, Dobbek, H. | | Deposit date: | 2014-06-25 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | Structural Basis for Organohalide Respiration.
Science, 346, 2014
|
|
4UWJ
 
 | | Crystal structure of Aspergillus fumigatus N-myristoyl transferase in complex with myristoyl CoA and a capped pyrazole sulphonamide ligand | | Descriptor: | 2,6-dichloro-N-(difluoromethyl)-4-[3-(piperidin-4-yl)propyl]-N-(1,3,5-trimethyl-1H-pyrazol-4-yl)benzenesulfonamide, GLYCYLPEPTIDE N-TETRADECANOYLTRANSFERASE, TETRADECANOYL-COA | | Authors: | Robinson, D.A, Brand, S, Norcross, N.R, Thompson, S, Harrison, J.R, Smith, V.C, Torrie, L.S, McElroy, S.P, Hallyburton, I, Norval, S, Stojanovski, L, Simeons, F.R.C, Frearson, J.A, Brenk, R, Fairlamb, A.H, Ferguson, M.A.J, Wyatt, P.G, Gilbert, I.H, Read, K.D. | | Deposit date: | 2014-08-12 | | Release date: | 2014-12-03 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Lead optimization of a pyrazole sulfonamide series of Trypanosoma brucei N-myristoyltransferase inhibitors: identification and evaluation of CNS penetrant compounds as potential treatments for stage 2 human African trypanosomiasis.
J. Med. Chem., 57, 2014
|
|
4UTI
 
 | | XenA - oxidized - Y183F variant in complex with coumarin | | Descriptor: | COUMARIN, FLAVIN MONONUCLEOTIDE, NADH:flavin oxidoreductase, ... | | Authors: | Werther, T, Dobbek, H. | | Deposit date: | 2014-07-21 | | Release date: | 2015-08-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.099 Å) | | Cite: | Redox-dependent substrate-cofactor interactions in the Michaelis-complex of a flavin-dependent oxidoreductase
Nat Commun, 8, 2017
|
|
7YSS
 
 | |
4UYG
 
 | | C-Terminal bromodomain of Human BRD2 with I-BET726 (GSK1324726A) | | Descriptor: | 4-[(2S,4R)-1-acetyl-4-[(4-chlorophenyl)amino]-2-methyl-1,2,3,4-tetrahydroquinolin-6-yl]benzoic acid, BROMODOMAIN-CONTAINING PROTEIN 2, SULFATE ION | | Authors: | Chung, C, Bamborough, P, Gosmini, R. | | Deposit date: | 2014-08-31 | | Release date: | 2014-10-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The Discovery of I-Bet726 (Gsk1324726A), a Potent Tetrahydroquinoline Apoa1 Up-Regulator and Selective Bet Bromodomain Inhibitor.
J.Med.Chem., 57, 2014
|
|
4UVJ
 
 | |
4UZ2
 
 | |
7YRW
 
 | |
4V01
 
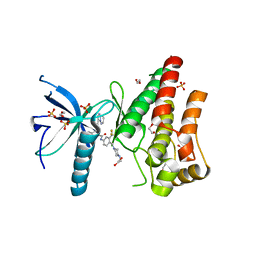 | | FGFR1 in complex with ponatinib (co-crystallisation). | | Descriptor: | 1,2-ETHANEDIOL, 3-(imidazo[1,2-b]pyridazin-3-ylethynyl)-4-methyl-N-{4-[(4-methylpiperazin-1-yl)methyl]-3-(trifluoromethyl)phenyl}benzam ide, FIBROBLAST GROWTH FACTOR RECEPTOR 1 (FMS-RELATED TYROSINE KINASE 2, ... | | Authors: | Tucker, J, Klein, T, Breed, J, Breeze, A, Overman, R, Phillips, C, Norman, R.A. | | Deposit date: | 2014-09-10 | | Release date: | 2014-12-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.33 Å) | | Cite: | Structural Insights Into Fgfr Kinase Isoform Selectivity: Diverse Binding Modes of Azd4547 and Ponatinib in Complex with Fgfr1 and Fgfr4
Structure, 22, 2014
|
|
6E8M
 
 | |
4V36
 
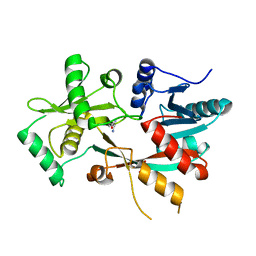 | | The structure of L-PGS from Bacillus licheniformis | | Descriptor: | 2,6-DIAMINO-HEXANOIC ACID AMIDE, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, LYSYL-TRNA-DEPENDENT L-YSYL-PHOSPHATIDYLGYCEROL SYNTHASE | | Authors: | Krausze, J, Hebecker, S, Heinz, D.W, Moser, J. | | Deposit date: | 2014-10-16 | | Release date: | 2015-08-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structures of Two Bacterial Resistance Factors Mediating tRNA-Dependent Aminoacylation of Phosphatidylglycerol with Lysine or Alanine.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4UZY
 
 | | Crystal structure of the Chlamydomonas IFT70 and IFT52 complex | | Descriptor: | CITRATE ANION, FLAGELLAR ASSOCIATED PROTEIN, INTRAFLAGELLAR TRANSPORT PROTEIN IFT52, ... | | Authors: | Taschner, M, Lorentzen, E. | | Deposit date: | 2014-09-09 | | Release date: | 2014-11-05 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.477 Å) | | Cite: | Crystal Structures of Ift70/52 and Ift52/46 Provide Insight Into Intraflagellar Transport B Core Complex Assembly.
J.Cell Biol., 207, 2014
|
|
6E5U
 
 | |
4V23
 
 | | RSV Matrix protein | | Descriptor: | MATRIX PROTEIN, POTASSIUM ION | | Authors: | Forster, A, Maertens, G, Bajorek, M. | | Deposit date: | 2014-10-05 | | Release date: | 2015-02-25 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Dimerization of Matrix Protein is Required for Budding of Respiratory Syncytial Virus.
J.Virol., 89, 2015
|
|
7YRE
 
 | | Crystal structure of a bright green fluorescent protein (StayGold) with triple mutations (N137A, Q140S, Y187F) in jellyfish Cytaeis uchidae from Biortus | | Descriptor: | 1,2-ETHANEDIOL, staygold(N137A,Q140S,Y187F) | | Authors: | Wu, J, Wang, F, Gui, W, Cheng, W, Yang, Y. | | Deposit date: | 2022-08-09 | | Release date: | 2023-08-16 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a bright green fluorescent protein (StayGold) with triple mutations (N137A, Q140S, Y187F) in jellyfish Cytaeis uchidae from Biortus
To Be Published
|
|
4V05
 
 | | FGFR1 in complex with AZD4547. | | Descriptor: | 1,2-ETHANEDIOL, FIBROBLAST GROWTH FACTOR RECEPTOR 1 (FMS-RELATED TYROSINE KINASE 2, PFEIFFER SYNDROME), ... | | Authors: | Tucker, J, Klein, T, Breed, J, Breeze, A, Overman, R, Phillips, C, Norman, R.A. | | Deposit date: | 2014-09-10 | | Release date: | 2014-12-10 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.57 Å) | | Cite: | Structural Insights Into Fgfr Kinase Isoform Selectivity: Diverse Binding Modes of Azd4547 and Ponatinib in Complex with Fgfr1 and Fgfr4
Structure, 22, 2014
|
|
4V3L
 
 | | RNF38-UB-UbcH5B-Ub complex | | Descriptor: | 1,2-ETHANEDIOL, E3 UBIQUITIN-PROTEIN LIGASE RNF38, POLYUBIQUITIN-C, ... | | Authors: | Buetow, L, Gabrielsen, M, Anthony, N.G, Dou, H, Patel, A, Aitkenhead, H, Sibbet, G.J, Smith, B.O, Huang, D.T. | | Deposit date: | 2014-10-20 | | Release date: | 2015-04-08 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Activation of a Primed Ring E3-E2-Ubiquitin Complex by Non-Covalent Ubiquitin.
Mol.Cell, 58, 2015
|
|
6E97
 
 | |
