5J18
 
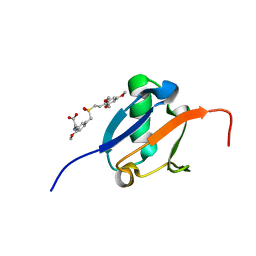 | | Solution structure of Ras Binding Domain (RBD) of B-Raf complexed with Rigosertib (Complex I) | | Descriptor: | N-[2-methoxy-5-({[(E)-2-(2,4,6-trimethoxyphenyl)ethenyl]sulfonyl}methyl)phenyl]glycine, Serine/threonine-protein kinase B-raf | | Authors: | Dutta, K, Vasquez-Del Carpio, R, Aggarwal, A.K, Reddy, E.P. | | Deposit date: | 2016-03-29 | | Release date: | 2016-05-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A Small Molecule RAS-Mimetic Disrupts RAS Association with Effector Proteins to Block Signaling.
Cell, 165, 2016
|
|
2RSO
 
 | |
2RTY
 
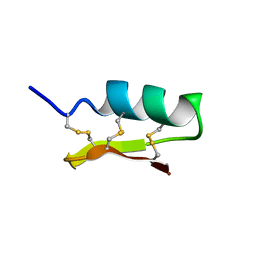 | | Solution structure of navitoxin | | Descriptor: | navitoxin | | Authors: | Umetsu, Y, Ohki, S. | | Deposit date: | 2013-10-16 | | Release date: | 2014-04-23 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Experimental conversion of a defensin into a neurotoxin: implications for origin of toxic function
MOL.BIOL.EVOL., 31, 2014
|
|
2ROL
 
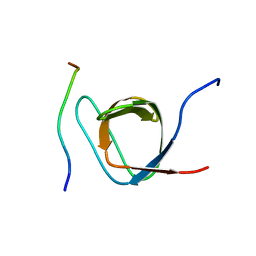 | | Structural Basis of PxxDY motif recognition in SH3 binding | | Descriptor: | 12-meric peptide from T-cell surface glycoprotein CD3 epsilon chain, Epidermal growth factor receptor kinase substrate 8-like protein 1 | | Authors: | Aitio, O, Hellman, M, Kesti, T, Kleino, I, Samuilova, O, Tossavainen, H, Paakkonen, K, Saksela, K, Permi, P. | | Deposit date: | 2008-04-02 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis of PxxDY motif recognition in SH3 binding
J.Mol.Biol., 382, 2008
|
|
2RTT
 
 | | Solution structure of the chitin-binding domain of Chi18aC from Streptomyces coelicolor | | Descriptor: | ChiC | | Authors: | Okumura, A, Uemura, M, Yamada, N, Chikaishi, E, Takai, T, Yoshio, S, Akagi, K, Morita, J, Lee, Y, Yokogawa, D, Suzuki, K, Watanabe, T, Ikegami, T. | | Deposit date: | 2013-08-26 | | Release date: | 2014-08-27 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution structure of the Chitin-binding domain of chitinase Chi18aC from Streptomyces coelicolor
To be Published
|
|
2RU0
 
 | |
2RON
 
 | | The external thioesterase of the Surfactin-Synthetase | | Descriptor: | Surfactin synthetase thioesterase subunit | | Authors: | Koglin, A, Lohr, F, Bernhard, F, Rogov, V.V, Frueh, D.P, Strieter, E.R, Mofid, M.R, Guentert, P, Wagner, G, Walsh, C.T, Marahiel, M.A, Doetsch, V. | | Deposit date: | 2008-04-04 | | Release date: | 2008-08-12 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Structural basis for the selectivity of the external thioesterase of the surfactin synthetase
Nature, 454, 2008
|
|
2RMN
 
 | |
2RMS
 
 | | Solution structure of the mSin3A PAH1-SAP25 SID complex | | Descriptor: | MSin3A-binding protein, Paired amphipathic helix protein Sin3a | | Authors: | Sahu, S.C, Swanson, K.A, Kang, R.S, Huang, K, Brubaker, K, Ratcliff, K, Radhakrishnan, I. | | Deposit date: | 2007-11-14 | | Release date: | 2008-01-22 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Conserved Themes in Target Recognition by the PAH1 and PAH2 Domains of the Sin3 Transcriptional Corepressor
J.Mol.Biol., 375, 2007
|
|
5TP6
 
 | |
5J2R
 
 | | Solution structure of Ras Binding Domain (RBD) of B-Raf | | Descriptor: | N-[2-methoxy-5-({[(E)-2-(2,4,6-trimethoxyphenyl)ethenyl]sulfonyl}methyl)phenyl]glycine, Serine/threonine-protein kinase B-raf | | Authors: | Dutta, K, Vasquez-Del Carpio, R, Aggarwal, A.K, Reddy, E.P. | | Deposit date: | 2016-03-29 | | Release date: | 2016-05-04 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | A Small Molecule RAS-Mimetic Disrupts RAS Association with Effector Proteins to Block Signaling.
Cell, 165, 2016
|
|
5J6U
 
 | |
5JPL
 
 | | LP2006, a handcuff-topology lasso peptide antibiotic | | Descriptor: | Uncharacterized protein | | Authors: | Tietz, J.I, Schwalen, C.J, Blair, P.M, Zakai, U.I, Mitchell, D.A. | | Deposit date: | 2016-05-03 | | Release date: | 2017-03-01 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | A new genome-mining tool redefines the lasso peptide biosynthetic landscape.
Nat. Chem. Biol., 13, 2017
|
|
6PQ2
 
 | |
5TTB
 
 | |
5TP5
 
 | |
6PMG
 
 | |
6PQE
 
 | |
6PRI
 
 | |
6PSI
 
 | |
5T56
 
 | |
5V4U
 
 | |
6QK5
 
 | |
2RSI
 
 | |
2RUG
 
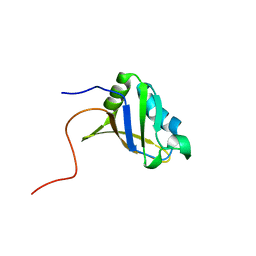 | | Refined solution structure of the first RNA recognition motif domain in CPEB3 | | Descriptor: | Cytoplasmic polyadenylation element-binding protein 3 | | Authors: | Tsuda, K, Kuwasako, K, Nagata, T, Takahashi, M, Kigawa, T, Kobayashi, N, Guntert, P, Shirouzu, M, Yokoyama, S, Muto, Y, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2014-04-15 | | Release date: | 2014-09-17 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Novel RNA recognition motif domain in the cytoplasmic polyadenylation element binding protein 3.
Proteins, 82, 2014
|
|
