4NIJ
 
 | | X-ray structure of the complex between hen egg white lysozyme and pentachlorocarbonyliridate(III) (30 days) | | Descriptor: | CHLORIDE ION, Lysozyme C, SODIUM ION, ... | | Authors: | Petruk, A.A, Bikiel, D.E, Vergara, A, Merlino, A. | | Deposit date: | 2013-11-06 | | Release date: | 2014-09-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Interaction between proteins and Ir based CO releasing molecules: mechanism of adduct formation and CO release.
Inorg.Chem., 53, 2014
|
|
4NIK
 
 | |
4NIL
 
 | |
4NIM
 
 | |
4NIN
 
 | |
4NIO
 
 | |
4NIP
 
 | |
4NIQ
 
 | | Crystal Structure of Vps4 MIT-Vfa1 MIM2 | | Descriptor: | VPS4-associated protein 1, Vacuolar protein sorting-associated protein 4 | | Authors: | Vild, C.J, Xu, Z. | | Deposit date: | 2013-11-06 | | Release date: | 2014-03-05 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Vfa1 Binds to the N-terminal Microtubule-interacting and Trafficking (MIT) Domain of Vps4 and Stimulates Its ATPase Activity.
J.Biol.Chem., 289, 2014
|
|
4NIR
 
 | |
4NIV
 
 | |
4NIW
 
 | |
4NIX
 
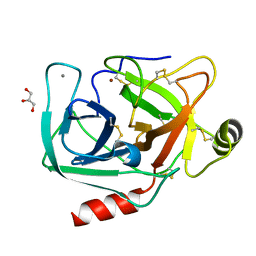 | | Crystal structure of trypsiligase (K60E/N143H/Y151H/D189K trypsin) orthorhombic form, zinc-bound | | Descriptor: | CALCIUM ION, Cationic trypsin, GLYCEROL, ... | | Authors: | Schoepfel, M, Parthier, C, Stubbs, M.T. | | Deposit date: | 2013-11-08 | | Release date: | 2014-02-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | N-terminal protein modification by substrate-activated reverse proteolysis.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4NIY
 
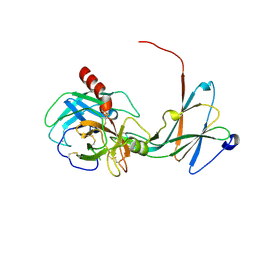 | | Crystal structure of trypsiligase (K60E/N143H/Y151H/D189K trypsin) complexed to YRH-ecotin (M84Y/M85R/A86H ecotin) | | Descriptor: | CALCIUM ION, Cationic trypsin, Ecotin, ... | | Authors: | Schoepfel, M, Parthier, C, Stubbs, M.T. | | Deposit date: | 2013-11-08 | | Release date: | 2014-02-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | N-terminal protein modification by substrate-activated reverse proteolysis.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
4NIZ
 
 | | GCN4-p1 single Val9 to aminobutyric acid mutant | | Descriptor: | GLYCEROL, General control protein GCN4 | | Authors: | Oshaben, K.M, Horne, W.S. | | Deposit date: | 2013-11-08 | | Release date: | 2014-08-20 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Tuning assembly size in Peptide-based supramolecular polymers by modulation of subunit association affinity.
Biomacromolecules, 15, 2014
|
|
4NJ0
 
 | | GCN4-p1 single Val9 to Ile mutant | | Descriptor: | General control protein GCN4 | | Authors: | Oshaben, K.M, Horne, W.S. | | Deposit date: | 2013-11-08 | | Release date: | 2014-08-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Tuning assembly size in Peptide-based supramolecular polymers by modulation of subunit association affinity.
Biomacromolecules, 15, 2014
|
|
4NJ1
 
 | | GCN4-p1 double Val9, 23 to Ile mutant | | Descriptor: | General control protein GCN4 | | Authors: | Oshaben, K.M, Horne, W.S. | | Deposit date: | 2013-11-08 | | Release date: | 2014-08-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Tuning assembly size in Peptide-based supramolecular polymers by modulation of subunit association affinity.
Biomacromolecules, 15, 2014
|
|
4NJ2
 
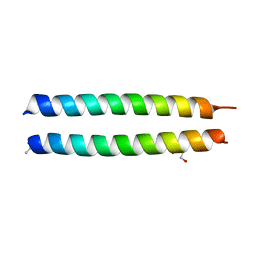 | | GCN4-p1 triple Val9, 23,30 to Ile mutant | | Descriptor: | GLYCEROL, General control protein GCN4 | | Authors: | Oshaben, K.M, Horne, W.S. | | Deposit date: | 2013-11-08 | | Release date: | 2014-08-20 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Tuning assembly size in Peptide-based supramolecular polymers by modulation of subunit association affinity.
Biomacromolecules, 15, 2014
|
|
4NJ3
 
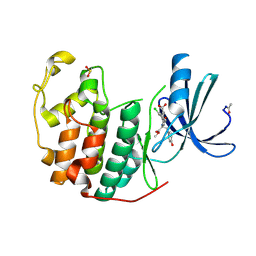 | |
4NJ4
 
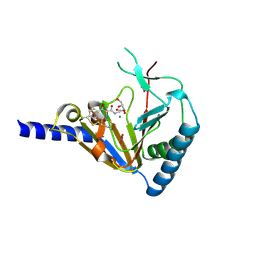 | | Crystal Structure of Human ALKBH5 | | Descriptor: | GLYCEROL, MANGANESE (II) ION, N-[(1-CHLORO-4-HYDROXYISOQUINOLIN-3-YL)CARBONYL]GLYCINE, ... | | Authors: | Aik, W.S, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2013-11-08 | | Release date: | 2014-01-15 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Structure of human RNA N6-methyladenine demethylase ALKBH5 provides insights into its mechanisms of nucleic acid recognition and demethylation.
Nucleic Acids Res., 42, 2014
|
|
4NJ5
 
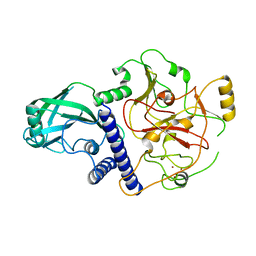 | | Crystal structure of SUVH9 | | Descriptor: | Probable histone-lysine N-methyltransferase, H3 lysine-9 specific SUVH9, ZINC ION | | Authors: | Du, J, Patel, D.J. | | Deposit date: | 2013-11-08 | | Release date: | 2014-01-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | SRA- and SET-domain-containing proteins link RNA polymerase V occupancy to DNA methylation.
Nature, 507, 2014
|
|
4NJ6
 
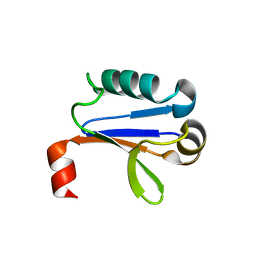 | | PB1 Domain of AtARF7 | | Descriptor: | Auxin response factor 7 | | Authors: | Korasick, D.A, Westfall, C.S, Lee, S.G, Nanao, M, Jez, J.M, Strader, L.C. | | Deposit date: | 2013-11-08 | | Release date: | 2014-03-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Molecular basis for AUXIN RESPONSE FACTOR protein interaction and the control of auxin response repression.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4NJ7
 
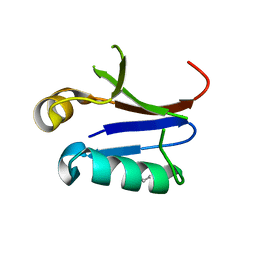 | | PB1 Domain of AtARF7 - SeMet Derivative | | Descriptor: | Auxin response factor 7 | | Authors: | Korasick, D.A, Westfall, C.S, Lee, S.G, Nanao, M, Jez, J.M, Strader, L.C. | | Deposit date: | 2013-11-08 | | Release date: | 2014-03-26 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Molecular basis for AUXIN RESPONSE FACTOR protein interaction and the control of auxin response repression.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
4NJ8
 
 | |
4NJ9
 
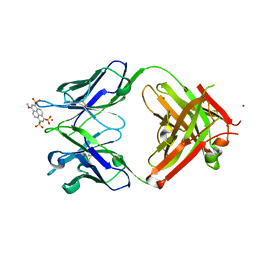 | | Crystal structure of Fab 8B10 in complex with MPTS | | Descriptor: | 8-methoxypyrene-1,3,6-trisulfonic acid, 8B10 heavy chain, 8B10 light chain, ... | | Authors: | Stanfield, R.L, Romesberg, F.E, Zimmermann, J, Wilson, I.A. | | Deposit date: | 2013-11-08 | | Release date: | 2014-11-12 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Adaptive Mutations Alter Antibody Structure and Dynamics during Affinity Maturation.
Biochemistry, 54, 2015
|
|
4NJA
 
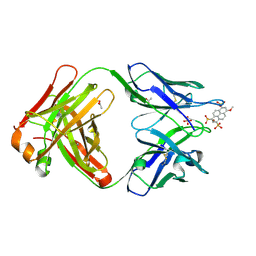 | | Crystal structure of Fab 6C8 in complex with MPTS | | Descriptor: | 6C8 heavy chain, 6C8 light chain, 8-methoxypyrene-1,3,6-trisulfonic acid, ... | | Authors: | Stanfield, R.L, Romesberg, F.E, Zimmermann, J, Wilson, I.A. | | Deposit date: | 2013-11-08 | | Release date: | 2014-11-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.204 Å) | | Cite: | Adaptive Mutations Alter Antibody Structure and Dynamics during Affinity Maturation.
Biochemistry, 54, 2015
|
|
