8H29
 
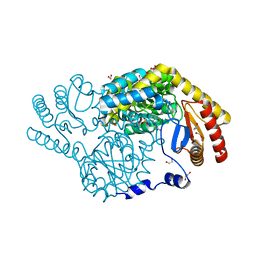 | | Serine Palmitoyltransferase from Sphingobacterium multivorum complexed with L-threonine | | Descriptor: | 1,2-ETHANEDIOL, N-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methyl)-L-threonine, Serine palmitoyltransferase | | Authors: | Murakami, T, Takahashi, A, Katayama, A, Miyahara, I, Kamiya, N, Ikushiro, H, Yano, T. | | Deposit date: | 2022-10-05 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural insights into the substrate recognition of serine palmitoyltransferase from Sphingobacterium multivorum.
J.Biol.Chem., 299, 2023
|
|
8HKF
 
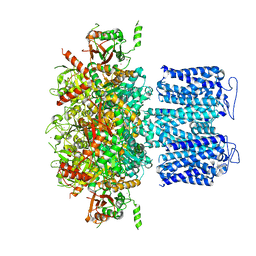 | | ion channel | | Descriptor: | POTASSIUM ION, Potassium channel subfamily T member 1, ZINC ION | | Authors: | Jiang, D.H, Zhang, Z.T. | | Deposit date: | 2022-11-25 | | Release date: | 2023-08-16 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Structural basis of human Slo2.2 channel gating and modulation.
Cell Rep, 42, 2023
|
|
8H1F
 
 | |
8H1E
 
 | |
8H1Y
 
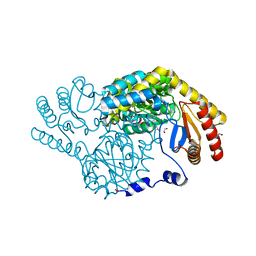 | | Serine Palmitoyltransferase from Sphingobacterium multivorum complexed with L-homoserine | | Descriptor: | (2~{S})-2-[[2-methyl-3-oxidanyl-5-(phosphonooxymethyl)pyridin-4-yl]methylamino]-4-oxidanyl-butanoic acid, 1,2-ETHANEDIOL, Serine palmitoyltransferase | | Authors: | Murakami, T, Takahashi, A, Katayama, A, Miyahara, I, Kamiya, N, Ikushiro, H, Yano, T. | | Deposit date: | 2022-10-04 | | Release date: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Structural insights into the substrate recognition of serine palmitoyltransferase from Sphingobacterium multivorum.
J.Biol.Chem., 299, 2023
|
|
7CL0
 
 | | Crystal structure of human SIRT6 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, NAD-dependent protein deacetylase sirtuin-6, ... | | Authors: | Song, K, Zhang, J. | | Deposit date: | 2020-07-20 | | Release date: | 2021-02-24 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.53 Å) | | Cite: | Reply to: Binding site for MDL-801 on SIRT6.
Nat.Chem.Biol., 17, 2021
|
|
2MEV
 
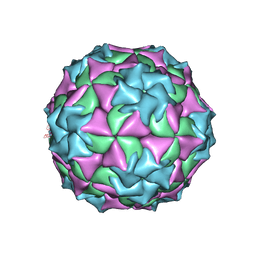 | | STRUCTURAL REFINEMENT AND ANALYSIS OF MENGO VIRUS | | Descriptor: | MENGO VIRUS COAT PROTEIN (SUBUNIT VP1), MENGO VIRUS COAT PROTEIN (SUBUNIT VP2), MENGO VIRUS COAT PROTEIN (SUBUNIT VP3), ... | | Authors: | Rossmann, M.G. | | Deposit date: | 1989-04-21 | | Release date: | 1990-01-15 | | Last modified: | 2023-04-19 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural refinement and analysis of Mengo virus.
J.Mol.Biol., 211, 1990
|
|
4UN0
 
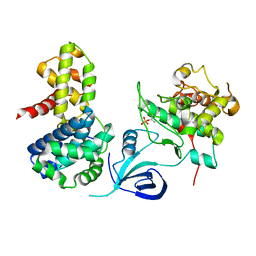 | | Crystal structure of the human CDK12-cyclinK complex | | Descriptor: | CYCLIN-DEPENDENT KINASE 12, CYCLIN-K | | Authors: | Dixon Clarke, S.E, Elkins, J.M, Pike, A.C.W, Chaikuad, A, Goubin, S, Krojer, T, Sorrell, F.J, Nowak, R, Williams, E, Kopec, J, Mahajan, R.P, Burgess-Brown, N, Carpenter, E.P, Knapp, S, von Delft, F, Arrowsmith, C.H, Edwards, A.M, Bountra, C, Bullock, A. | | Deposit date: | 2014-05-22 | | Release date: | 2014-06-04 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Structures of the Cdk12/Cyck Complex with AMP-Pnp Reveal a Flexible C-Terminal Kinase Extension Important for ATP Binding.
Sci.Rep., 5, 2015
|
|
8H0H
 
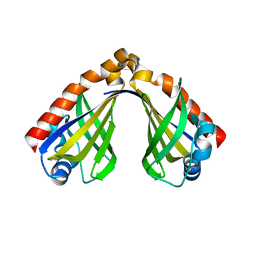 | |
8H5B
 
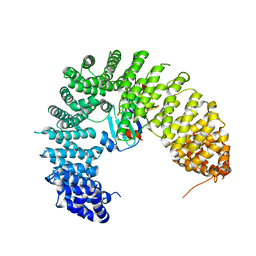 | | The cryo-EM structure of nuclear transport receptor Kap114p complex with yeast TATA-box binding protein | | Descriptor: | Importin subunit beta-5, TATA-box-binding protein | | Authors: | Hsia, K.C, Liao, C.C, Wang, C.H, Wu, Y.M. | | Deposit date: | 2022-10-12 | | Release date: | 2023-09-20 | | Method: | ELECTRON MICROSCOPY (4.03 Å) | | Cite: | Structural convergence endows nuclear transport receptor Kap114p with a transcriptional repressor function toward TATA-binding protein.
Nat Commun, 14, 2023
|
|
8HEA
 
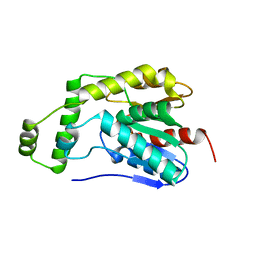 | | Esterase2 (EaEst2) from Exiguobacterium antarcticum | | Descriptor: | Thermostable carboxylesterase Est30 | | Authors: | Hwang, J, Lee, J.H. | | Deposit date: | 2022-11-08 | | Release date: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural and Biochemical Insights into Bis(2-hydroxyethyl) Terephthalate Degrading Carboxylesterase Isolated from Psychrotrophic Bacterium Exiguobacterium antarcticum.
Int J Mol Sci, 24, 2023
|
|
5A87
 
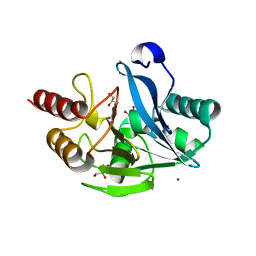 | | Crystal structure of the metallo-beta-lactamase VIM-5 | | Descriptor: | GLYCEROL, METALLO-BETA-LACTAMASE VIM-5, ZINC ION | | Authors: | Brem, J, Duzgun, A.O, McDonough, M.A, Schofield, C.J. | | Deposit date: | 2015-07-13 | | Release date: | 2015-12-23 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Comparison of Verona Integron-Borne Metallo-beta-Lactamase (VIM) Variants Reveals Differences in Stability and Inhibition Profiles.
Antimicrob. Agents Chemother., 60, 2015
|
|
2MEJ
 
 | |
4V7F
 
 | | Arx1 pre-60S particle. | | Descriptor: | 25S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Leidig, C, Thoms, M, Holdermann, I, Bradatsch, B, Berninghausen, O, Bange, G, Sinning, I, Hurt, E, Beckmann, R. | | Deposit date: | 2013-12-10 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (8.7 Å) | | Cite: | 60S ribosome biogenesis requires rotation of the 5S ribonucleoprotein particle.
Nat Commun, 5, 2014
|
|
4UY2
 
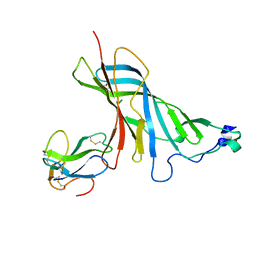 | | Crystal structure of the complex of the extracellular domain of human alpha9 nAChR with alpha-bungarotoxin. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ALPHA-BUNGAROTOXIN ISOFORM V31, NEURONAL ACETYLCHOLINE RECEPTOR SUBUNIT ALPHA-9 | | Authors: | Giastas, P, Zouridakis, M, Zarkadas, E, Tzartos, S.J. | | Deposit date: | 2014-08-28 | | Release date: | 2014-10-01 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.697 Å) | | Cite: | Crystal Structures of Free and Antagonist-Bound States of Human Alpha9 Nicotinic Receptor Extracellular Domain
Nat.Struct.Mol.Biol., 21, 2014
|
|
2LZM
 
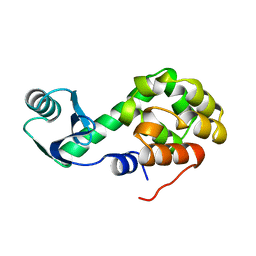 | |
4V1Z
 
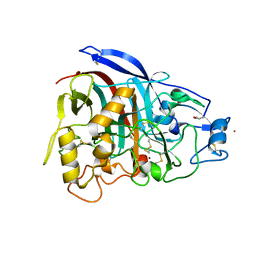 | | The 3-D structure of the cellobiohydrolase, Cel7A, from Aspergillus fumigatus | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CELLOBIOHYDROLASE, ZINC ION | | Authors: | Moroz, O.V, Maranta, M, Shaghasi, T, Harris, P.V, Wilson, K.S, Davies, G.J. | | Deposit date: | 2014-10-04 | | Release date: | 2015-01-14 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | The Three-Dimensional Structure of the Cellobiohydrolase Cel7A from Aspergillus Fumigatus at 1.5 A Resolution
Acta Crystallogr.,Sect.F, 71, 2015
|
|
4V48
 
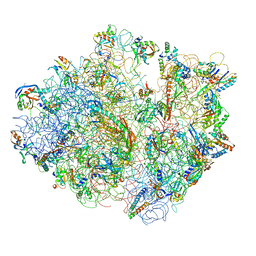 | | Real space refined coordinates of the 30S and 50S subunits fitted into the low resolution cryo-EM map of the initiation-like state of E. coli 70S ribosome | | Descriptor: | 16S RIBOSOMAL RNA, 23S ribosomal RNA, 30S RIBOSOMAL PROTEIN S10, ... | | Authors: | Gao, H, Sengupta, J, Valle, M, Korostelev, A, Eswar, N, Stagg, S.M, Van Roey, P, Agrawal, R.K, Harvey, S.T, Sali, A, Chapman, M.S, Frank, J. | | Deposit date: | 2003-05-06 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (11.5 Å) | | Cite: | Study of the structural dynamics of the E. coli 70S ribosome using real space refinement
Cell(Cambridge,Mass.), 113, 2003
|
|
4V6Q
 
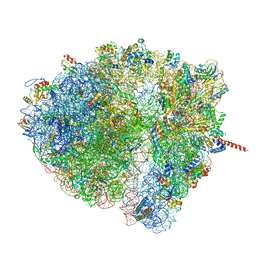 | | Structural characterization of mRNA-tRNA translocation intermediates (class 5 of the six classes) | | Descriptor: | 16S ribosomal RNA, 23S ribosomal RNA, 30S ribosomal protein S10, ... | | Authors: | Agirrezabala, X, Liao, H, Schreiner, E, Fu, J, Ortiz-Meoz, R.F, Schulten, K, Green, R, Frank, J. | | Deposit date: | 2011-12-08 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (11.5 Å) | | Cite: | Structural characterization of mRNA-tRNA translocation intermediates.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4V7H
 
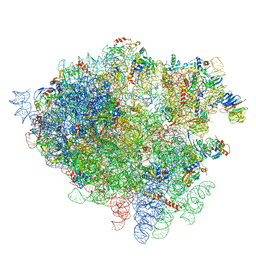 | | Structure of the 80S rRNA and proteins and P/E tRNA for eukaryotic ribosome based on cryo-EM map of Thermomyces lanuginosus ribosome at 8.9A resolution | | Descriptor: | 18S rRNA, 26S ribosomal RNA, 40S ribosomal protein S0(A), ... | | Authors: | Taylor, D.J, Devkota, B, Huang, A.D, Topf, M, Narayanan, E, Sali, A, Harvey, S.C, Frank, J. | | Deposit date: | 2009-09-22 | | Release date: | 2014-07-09 | | Last modified: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (8.9 Å) | | Cite: | Comprehensive molecular structure of the eukaryotic ribosome.
Structure, 17, 2009
|
|
4V8Z
 
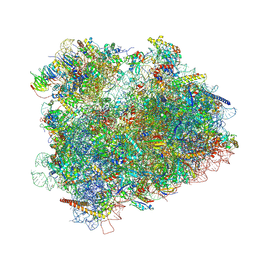 | | Cryo-EM reconstruction of the 80S-eIF5B-Met-itRNAMet Eukaryotic Translation Initiation Complex | | Descriptor: | 18S RIBOSOMAL RNA, 25S RIBOSOMAL RNA, 40S RIBOSOMAL PROTEIN S0-A, ... | | Authors: | Fernandez, I.S, Bai, X.C, Hussain, T, Kelley, A.C, Lorsch, J.R, Ramakrishnan, V, Scheres, S.H.W. | | Deposit date: | 2013-07-20 | | Release date: | 2014-07-09 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (6.6 Å) | | Cite: | Molecular architecture of a eukaryotic translational initiation complex.
Science, 342, 2013
|
|
4WB7
 
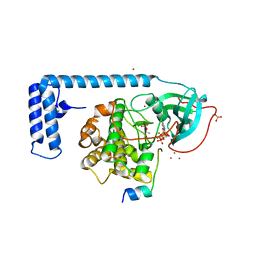 | | Crystal structure of a chimeric fusion of human DnaJ (Hsp40) and cAMP-dependent protein kinase A (catalytic alpha subunit) | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, DnaJ homolog subfamily B member 1,cAMP-dependent protein kinase catalytic subunit alpha, PKI (5-24), ... | | Authors: | Cheung, J, Ginter, C, Cassidy, M, Franklin, M.C, Rudolph, M.J, Hendrickson, W.A. | | Deposit date: | 2014-09-02 | | Release date: | 2015-01-21 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural insights into mis-regulation of protein kinase A in human tumors.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4WBU
 
 | | prion peptide | | Descriptor: | PrP peptide | | Authors: | Yu, L, Lee, S.-J, Yee, V. | | Deposit date: | 2014-09-03 | | Release date: | 2015-05-27 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Crystal Structures of Polymorphic Prion Protein beta 1 Peptides Reveal Variable Steric Zipper Conformations.
Biochemistry, 54, 2015
|
|
4V6W
 
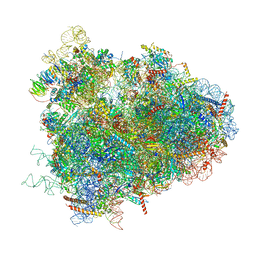 | | Structure of the D. melanogaster 80S ribosome | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA, 2S ribosomal RNA, ... | | Authors: | Anger, A.M, Armache, J.-P, Berninghausen, O, Habeck, M, Subklewe, M, Wilson, D.N, Beckmann, R. | | Deposit date: | 2013-02-27 | | Release date: | 2014-07-09 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | Structures of the human and Drosophila 80S ribosome.
Nature, 497, 2013
|
|
4V7N
 
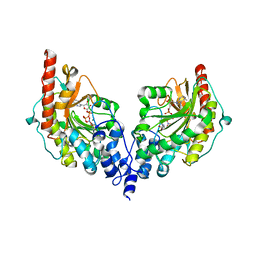 | | Glycocyamine kinase, beta-beta homodimer from marine worm Namalycastis sp., with transition state analog Mg(II)-ADP-NO3-glycocyamine. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, GUANIDINO ACETATE, Glycocyamine kinase beta chain, ... | | Authors: | Lim, K, Pullalarevu, S, Herzberg, O. | | Deposit date: | 2009-12-15 | | Release date: | 2014-07-09 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for the mechanism and substrate specificity of glycocyamine kinase, a phosphagen kinase family member.
Biochemistry, 49, 2010
|
|
