1EZ4
 
 | | CRYSTAL STRUCTURE OF NON-ALLOSTERIC L-LACTATE DEHYDROGENASE FROM LACTOBACILLUS PENTOSUS AT 2.3 ANGSTROM RESOLUTION | | Descriptor: | LACTATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Uchikoba, H, Fushinobu, S, Wakagi, T, Konno, M, Taguchi, H, Matsuzawa, H. | | Deposit date: | 2000-05-10 | | Release date: | 2001-12-28 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of non-allosteric L-lactate dehydrogenase from Lactobacillus pentosus at 2.3 A resolution: specific interactions at subunit interfaces.
Proteins, 46, 2002
|
|
8LDH
 
 | |
8XKV
 
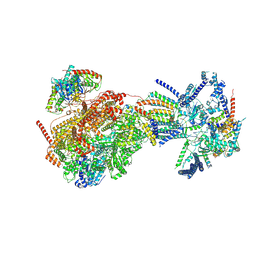 | | Cryo-EM structure of the Ycf2-FtsHi motor complex from Arabidopsis in Apo state | | Descriptor: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, ATP-dependent zinc metalloprotease FTSH 12, chloroplastic, ... | | Authors: | Liang, K, Zhan, X, Xu, Q, Wu, J, Yan, Z. | | Deposit date: | 2023-12-25 | | Release date: | 2024-09-11 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Structural insights into the chloroplast protein import in land plants.
Cell, 187, 2024
|
|
8XKU
 
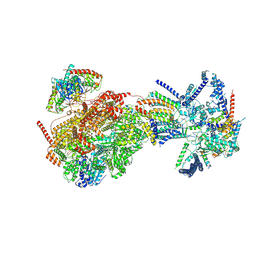 | | Cryo-EM structure of the Ycf2-FtsHi motor complex from Arabidopsis in ATP-bound state | | Descriptor: | 1,2-DILAUROYL-SN-GLYCERO-3-PHOSPHATE, ADENOSINE-5'-TRIPHOSPHATE, ATP-dependent zinc metalloprotease FTSH 12, ... | | Authors: | Liang, K, Zhan, X, Xu, Q, Wu, J, Yan, Z. | | Deposit date: | 2023-12-24 | | Release date: | 2024-09-11 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural insights into the chloroplast protein import in land plants.
Cell, 187, 2024
|
|
8Q3C
 
 | | Structure of Selenomonas ruminantium lactate dehydrogenase I85R mutant | | Descriptor: | CHLORIDE ION, L-lactate dehydrogenase, NITRATE ION, ... | | Authors: | Bertrand, Q, Coquille, S, Iorio, A, Sterpone, F, Madern, D. | | Deposit date: | 2023-08-03 | | Release date: | 2023-11-01 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Biochemical, structural and dynamical characterizations of the lactate dehydrogenase from Selenomonas ruminantium provide information about an intermediate evolutionary step prior to complete allosteric regulation acquisition in the super family of lactate and malate dehydrogenases.
J.Struct.Biol., 215, 2023
|
|
6OR9
 
 | |
8J5D
 
 | | Cryo-EM structure of starch degradation complex of BAM1-LSF1-MDH | | Descriptor: | Beta-amylase 1, chloroplastic, Malate dehydrogenase, ... | | Authors: | Guan, Z.Y, Liu, J, Yan, J.J. | | Deposit date: | 2023-04-21 | | Release date: | 2024-01-10 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | The LIKE SEX FOUR 1-malate dehydrogenase complex functions as a scaffold to recruit beta-amylase to promote starch degradation.
Plant Cell, 36, 2023
|
|
3NEP
 
 | |
2PWZ
 
 | |
4PLT
 
 | | Crystal structure of ancestral apicomplexan malate dehydrogenase with oxamate. | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, OXAMIC ACID, malate dehydrogenase | | Authors: | Boucher, J.I, Jacobowitz, J.R, Beckett, B.C, Classen, S, Theobald, D.L. | | Deposit date: | 2014-05-19 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | An atomic-resolution view of neofunctionalization in the evolution of apicomplexan lactate dehydrogenases.
Elife, 3, 2014
|
|
4PLF
 
 | | Crystal structure of ancestral apicomplexan lactate dehydrogenase with lactate. | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PYRUVIC ACID, ... | | Authors: | Boucher, J.I, Jacobowitz, J.R, Beckett, B.C, Classen, S, Theobald, D.L. | | Deposit date: | 2014-05-16 | | Release date: | 2014-07-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | An atomic-resolution view of neofunctionalization in the evolution of apicomplexan lactate dehydrogenases.
Elife, 3, 2014
|
|
4PLZ
 
 | | Crystal structure of Plasmodium falciparum lactate dehydrogenase mutant W107fA. | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, L-lactate dehydrogenase, OXAMIC ACID | | Authors: | Boucher, J.I, Jacobowitz, J.R, Beckett, B.C, Classen, S, Theobald, D.L. | | Deposit date: | 2014-05-20 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.05 Å) | | Cite: | An atomic-resolution view of neofunctionalization in the evolution of apicomplexan lactate dehydrogenases.
Elife, 3, 2014
|
|
4PLG
 
 | | Crystal structure of ancestral apicomplexan lactate dehydrogenase with oxamate. | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, OXAMIC ACID, SODIUM ION, ... | | Authors: | Boucher, J.I, Jacobowitz, J.R, Beckett, B.C, Classen, S, Theobald, D.L. | | Deposit date: | 2014-05-16 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.192 Å) | | Cite: | An atomic-resolution view of neofunctionalization in the evolution of apicomplexan lactate dehydrogenases.
Elife, 3, 2014
|
|
4PLY
 
 | | Crystal structure of ancestral apicomplexan malate dehydrogenase with malate. | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, PYRUVIC ACID, malate dehydrogenase | | Authors: | Boucher, J.I, Jacobowitz, J.R, Beckett, B.C, Classen, S, Theobald, D.L. | | Deposit date: | 2014-05-19 | | Release date: | 2014-07-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | An atomic-resolution view of neofunctionalization in the evolution of apicomplexan lactate dehydrogenases.
Elife, 3, 2014
|
|
4Q3N
 
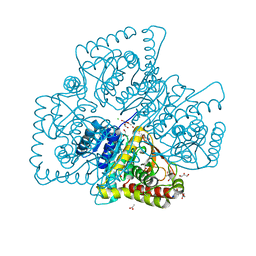 | | Crystal structure of MGS-M5, a lactate dehydrogenase enzyme from a Medee basin deep-sea metagenome library | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ACETATE ION, CHLORIDE ION, ... | | Authors: | Stogios, P.J, Xu, X, Cui, H, Alcaide, M, Ferrer, M, Savchenko, A. | | Deposit date: | 2014-04-11 | | Release date: | 2015-02-25 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | Pressure adaptation is linked to thermal adaptation in salt-saturated marine habitats.
Environ Microbiol, 17, 2015
|
|
4PLC
 
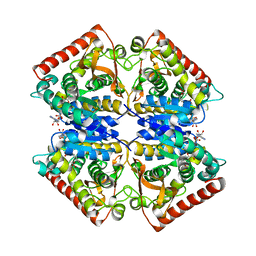 | | Crystal structure of ancestral apicomplexan lactate dehydrogenase with malate. | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, PYRUVIC ACID, ... | | Authors: | Boucher, J.I, Jacobowitz, J.R, Beckett, B.C, Classen, S, Theobald, D.L. | | Deposit date: | 2014-05-16 | | Release date: | 2014-07-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | An atomic-resolution view of neofunctionalization in the evolution of apicomplexan lactate dehydrogenases.
Elife, 3, 2014
|
|
4PLW
 
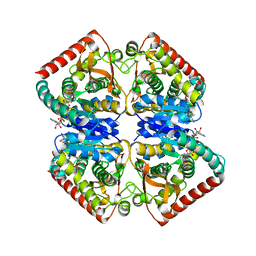 | | Crystal structure of ancestral apicomplexan malate dehydrogenase with lactate. | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, PHOSPHATE ION, malate dehydrogenase | | Authors: | Boucher, J.I, Jacobowitz, J.R, Beckett, B.C, Classen, S, Theobald, D.L. | | Deposit date: | 2014-05-19 | | Release date: | 2014-07-02 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | An atomic-resolution view of neofunctionalization in the evolution of apicomplexan lactate dehydrogenases.
Elife, 3, 2014
|
|
4PLV
 
 | | Crystal structure of ancestral apicomplexan malate dehydrogenase with lactate. | | Descriptor: | (2S)-2-HYDROXYPROPANOIC ACID, 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, malate dehydrogenase | | Authors: | Boucher, J.I, Jacobowitz, J.R, Beckett, B.C, Classen, S, Theobald, D.L. | | Deposit date: | 2014-05-19 | | Release date: | 2014-07-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | An atomic-resolution view of neofunctionalization in the evolution of apicomplexan lactate dehydrogenases.
Elife, 3, 2014
|
|
9QCG
 
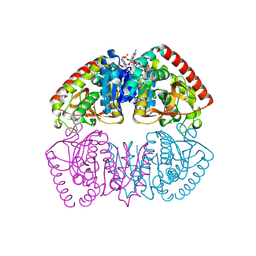 | |
4L4S
 
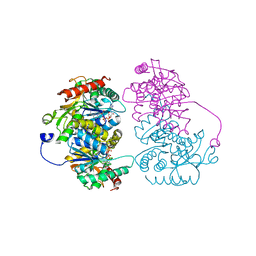 | | Structural characterisation of the NADH binary complex of human lactate dehydrogenase M isozyme | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, L-lactate dehydrogenase A chain | | Authors: | Dempster, S, Harper, S, Moses, J.E, Dreveny, I. | | Deposit date: | 2013-06-09 | | Release date: | 2014-04-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural characterization of the apo form and NADH binary complex of human lactate dehydrogenase.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
9LDT
 
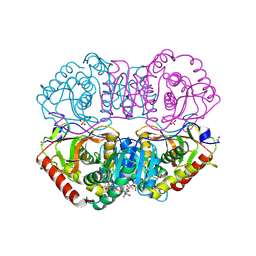 | | DESIGN AND SYNTHESIS OF NEW ENZYMES BASED ON THE LACTATE DEHYDROGENASE FRAMEWORK | | Descriptor: | LACTATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, OXAMIC ACID, ... | | Authors: | Dunn, C.R, Holbrook, J.J, Muirhead, H. | | Deposit date: | 1991-11-26 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Design and synthesis of new enzymes based on the lactate dehydrogenase framework.
Philos.Trans.R.Soc.London,Ser.B, 332, 1991
|
|
9LDB
 
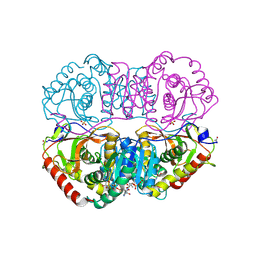 | | DESIGN AND SYNTHESIS OF NEW ENZYMES BASED ON THE LACTATE DEHYDROGENASE FRAMEWORK | | Descriptor: | LACTATE DEHYDROGENASE, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, OXAMIC ACID, ... | | Authors: | Dunn, C.R, Holbrook, J.J, Muirhead, H. | | Deposit date: | 1991-11-26 | | Release date: | 1993-10-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Design and synthesis of new enzymes based on the lactate dehydrogenase framework.
Philos.Trans.R.Soc.London,Ser.B, 332, 1991
|
|
8RS5
 
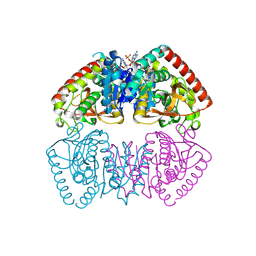 | | Crystal structure of Methanopyrus kandleri malate dehydrogenase mutant 4 | | Descriptor: | CHLORIDE ION, Malate dehydrogenase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, ... | | Authors: | Coquille, S, Engilberge, S, Madern, D. | | Deposit date: | 2024-01-24 | | Release date: | 2024-07-10 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Allostery and Evolution: A Molecular Journey Through the Structural and Dynamical Landscape of an Enzyme Super Family.
Mol.Biol.Evol., 42, 2025
|
|
8RWL
 
 | | Crystal structure of Methanopyrus kandleri malate dehydrogenase mutant 1 | | Descriptor: | CHLORIDE ION, GLYCEROL, Malate dehydrogenase, ... | | Authors: | Coquille, S, Roche, J, Engilberge, S, Girard, E, Madern, D. | | Deposit date: | 2024-02-05 | | Release date: | 2024-07-10 | | Last modified: | 2025-01-29 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Allostery and Evolution: A Molecular Journey Through the Structural and Dynamical Landscape of an Enzyme Super Family.
Mol.Biol.Evol., 42, 2025
|
|
4LN1
 
 | | CRYSTAL STRUCTURE OF L-lactate dehydrogenase from Bacillus cereus ATCC 14579 complexed with calcium, NYSGRC Target 029452 | | Descriptor: | CALCIUM ION, L-lactate dehydrogenase 1 | | Authors: | Malashkevich, V.N, Bonanno, J.B, Bhosle, R, Toro, R, Hillerich, B, Gizzi, A, Garforth, S, Kar, A, Chan, M.K, Lafluer, J, Patel, H, Matikainen, B, Chamala, S, Lim, S, Celikgil, A, Villegas, G, Evans, B, Love, J, Fiser, A, Khafizov, K, Seidel, R, Almo, S.C, New York Structural Genomics Research Consortium (NYSGRC) | | Deposit date: | 2013-07-11 | | Release date: | 2013-07-24 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of L-lactate dehydrogenase from Bacillus cereus ATCC 14579 complexed with calcium, NYSGRC Target 029452
To be Published
|
|
