1BAV
 
 | |
6UZY
 
 | | Cryo-EM structure of Xenopus tropicalis pannexin 1 | | Descriptor: | HEXADECANE, Pannexin, [(2~{R})-1-[2-azanylethoxy(oxidanyl)phosphoryl]oxy-3-hexadecanoyloxy-propan-2-yl] (~{Z})-octadec-9-enoate | | Authors: | Deng, Z, He, Z, Yuan, P. | | Deposit date: | 2019-11-15 | | Release date: | 2020-04-01 | | Last modified: | 2020-04-22 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | Cryo-EM structures of the ATP release channel pannexin 1.
Nat.Struct.Mol.Biol., 27, 2020
|
|
6NBP
 
 | | Crystal Structure of a Sugar N-Formyltransferase from the Plant Pathogen Pantoea ananatis | | Descriptor: | N-formyltransferase, N-{[4-({[(6R)-2-amino-5-formyl-4-oxo-1,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)phenyl]carbonyl}-L-glutamic acid, PHOSPHATE ION, ... | | Authors: | Hofmeister, D.L, Thoden, J.B, Holden, H.M. | | Deposit date: | 2018-12-09 | | Release date: | 2018-12-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Investigation of a sugar N-formyltransferase from the plant pathogen Pantoea ananatis.
Protein Sci., 28, 2019
|
|
1C8F
 
 | | FELINE PANLEUKOPENIA VIRUS EMPTY CAPSID STRUCTURE | | Descriptor: | CALCIUM ION, FELINE PANLEUKOPENIA VIRUS CAPSID | | Authors: | Rossmann, M.G, Simpson, A.A. | | Deposit date: | 2000-05-05 | | Release date: | 2000-08-09 | | Last modified: | 2023-04-19 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Host range and variability of calcium binding by surface loops in the capsids of canine and feline parvoviruses.
J.Mol.Biol., 300, 2000
|
|
1C8G
 
 | | FELINE PANLEUKOPENIA VIRUS EMPTY CAPSID STRUCTURE | | Descriptor: | CALCIUM ION, FELINE PANLEUKOPENIA VIRUS CAPSID | | Authors: | Rossmann, M.G, Simpson, A.A. | | Deposit date: | 2000-05-05 | | Release date: | 2000-08-09 | | Last modified: | 2023-04-19 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Host range and variability of calcium binding by surface loops in the capsids of canine and feline parvoviruses.
J.Mol.Biol., 300, 2000
|
|
5HG0
 
 | | Crystal Structure of Pantoate-beta-alanine Ligase from Francisella tularensis complex with SAM | | Descriptor: | Pantothenate synthetase, S-ADENOSYLMETHIONINE | | Authors: | Chang, C, Maltseva, N, Kim, Y, Papazisi, L, Anderson, W.F, Joachimiak, A, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2016-01-07 | | Release date: | 2016-01-20 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal Structure of Pantoate-beta-alanine Ligase from Francisella tularensis complex with SAM
To Be Published
|
|
6Z7C
 
 | | Variant Surface Glycoprotein VSGsur mutant H122A | | Descriptor: | Variant surface glycoprotein Sur, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Zeelen, J.P, Straaten van, M, Stebbins, C.E. | | Deposit date: | 2020-05-30 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.64 Å) | | Cite: | Structure of trypanosome coat protein VSGsur and function in suramin resistance.
Nat Microbiol, 6, 2021
|
|
6Z7E
 
 | |
6Z7B
 
 | | Variant Surface Glycoprotein VSGsur bound to suramin | | Descriptor: | 8,8'-[CARBONYLBIS[IMINO-3,1-PHENYLENECARBONYLIMINO(4-METHYL-3,1-PHENYLENE)CARBONYLIMINO]]BIS-1,3,5-NAPHTHALENETRISULFON IC ACID, Variant surface glycoprotein Sur, alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Zeelen, J.P, Straaten van, M, Stebbins, C.E, Jeffrey, P. | | Deposit date: | 2020-05-30 | | Release date: | 2021-03-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Structure of trypanosome coat protein VSGsur and function in suramin resistance.
Nat Microbiol, 6, 2021
|
|
6Z7D
 
 | | Variant Surface Glycoprotein VSGsur mutant H122A soaked in 0.77 mM Suramin. | | Descriptor: | Variant surface glycoprotein Sur, alpha-D-mannopyranose-(1-3)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Zeelen, J.P, Straaten van, M, Stebbins, C.E. | | Deposit date: | 2020-05-30 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of trypanosome coat protein VSGsur and function in suramin resistance.
Nat Microbiol, 6, 2021
|
|
6Z79
 
 | | Variant Surface Glycoprotein VSGsur, I3C ("Magic Triangle") derivative used for phasing of the structure and subsequently as a model for molecular replacement of native, mutants, and drug soaks. | | Descriptor: | 5-amino-2,4,6-triiodobenzene-1,3-dicarboxylic acid, Variant surface glycoprotein Sur, alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Zeelen, J.P, Straaten van, M, Stebbins, C.E. | | Deposit date: | 2020-05-30 | | Release date: | 2021-03-17 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Structure of trypanosome coat protein VSGsur and function in suramin resistance.
Nat Microbiol, 6, 2021
|
|
5I2G
 
 | | 1,2-propanediol Dehydration in Roseburia inulinivorans; Structural Basis for Substrate and Enantiomer Selectivity | | Descriptor: | Diol dehydratase, S-1,2-PROPANEDIOL | | Authors: | LaMattina, J.W, Reitzer, P, Kapoor, S, Galzerani, F, Koch, D.J, Gouvea, I.E, Lanzilotta, W.N. | | Deposit date: | 2016-02-08 | | Release date: | 2016-06-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.352 Å) | | Cite: | 1,2-Propanediol Dehydration in Roseburia inulinivorans: STRUCTURAL BASIS FOR SUBSTRATE AND ENANTIOMER SELECTIVITY.
J.Biol.Chem., 291, 2016
|
|
6NIM
 
 | | Trypanosoma cruzi - BDF2, TcCLB.506553.20, solved with bromosporine | | Descriptor: | Bromodomain factor 2 protein, Bromosporine, SULFATE ION, ... | | Authors: | Lin, Y.H, Dong, A, Tempel, W, Loppnau, P, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Hui, R, Vedadi, M, Harding, R, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-29 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Trypanosoma cruzi - BDF2, TcCLB.506553.20, solved with bromosporine
to be published
|
|
8RTH
 
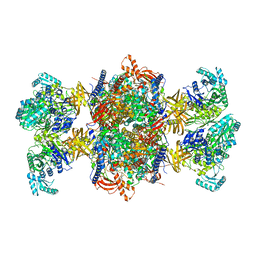 | | Trypanosoma brucei 3-methylcrotonyl-CoA carboxylase | | Descriptor: | 3-methylcrotonyl-CoA carboxylase, putative, 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, ... | | Authors: | Ruiz, F.M, Plaza-Pegueroles, A, Fernandez-Tornero, C. | | Deposit date: | 2024-01-26 | | Release date: | 2024-04-17 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.37 Å) | | Cite: | The cryo-EM structure of trypanosome 3-methylcrotonyl-CoA carboxylase provides mechanistic and dynamic insights into its enzymatic function.
Structure, 2024
|
|
6NEZ
 
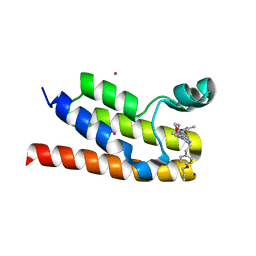 | | Trypanosoma brucei - BDF5, Tb427tmp.01.5000 A, solved with PF-CBP1 | | Descriptor: | 5-(3,5-dimethyl-1,2-oxazol-4-yl)-1-[2-(morpholin-4-yl)ethyl]-2-[2-(4-propoxyphenyl)ethyl]-1H-benzimidazole, UNKNOWN ATOM OR ION, Uncharacterized protein | | Authors: | Lin, Y.H, Dong, A, Tempel, W, McAuley, J, Loppnau, P, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Hui, R, Vedadi, M, Harding, R.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2018-12-18 | | Release date: | 2019-01-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Trypanosoma brucei - BDF5, Tb427tmp.01.5000 A, solved with PF-CBP1
to be published
|
|
2HCZ
 
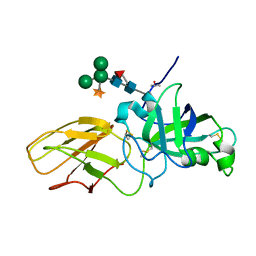 | | Crystal structure of EXPB1 (Zea m 1), a beta-expansin and group-1 pollen allergen from maize | | Descriptor: | Beta-expansin 1a, alpha-D-xylopyranose-(1-2)-[alpha-D-mannopyranose-(1-3)][alpha-D-mannopyranose-(1-6)]alpha-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-D-fucopyranose-(1-3)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Yennawar, N.H, Cosgrove, D.J. | | Deposit date: | 2006-06-19 | | Release date: | 2006-08-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Crystal structure and activities of EXPB1 (Zea m 1), a beta-expansin and group-1 pollen allergen from maize.
Proc.Natl.Acad.Sci.Usa, 103, 2006
|
|
2IIT
 
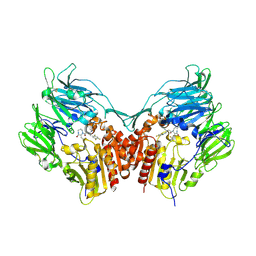 | | Human dipeptidyl peptidase 4 in complex with a diazepan-2-one inhibitor | | Descriptor: | (3R)-4-[(3R)-3-AMINO-4-(2,4,5-TRIFLUOROPHENYL)BUTANOYL]-3-(2,2,2-TRIFLUOROETHYL)-1,4-DIAZEPAN-2-ONE, 2-acetamido-2-deoxy-alpha-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Scapin, G, Biftu, T, Weber, A.E. | | Deposit date: | 2006-09-28 | | Release date: | 2006-11-28 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | (3R)-4-[(3R)-3-Amino-4-(2,4,5-trifluorophenyl)butanoyl]-3-(2,2,2-trifluoroethyl)-1,4-diazepan-2-one, a selective dipeptidyl peptidase IV inhibitor for the treatment of type 2 diabetes
Bioorg.Med.Chem.Lett., 17, 2007
|
|
5V6S
 
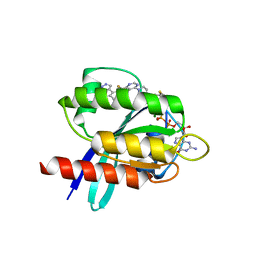 | | Crystal structure of small molecule acrylamide 1 covalently bound to K-Ras G12C | | Descriptor: | 1-{4-[6-chloro-8-fluoro-7-(5-methyl-1H-indazol-4-yl)quinazolin-4-yl]piperazin-1-yl}propan-1-one, GTPase KRas, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | McGregor, L.M, Jenkins, M, Kerwin, C, Burke, J.E, Shokat, K.M. | | Deposit date: | 2017-03-17 | | Release date: | 2017-06-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Expanding the Scope of Electrophiles Capable of Targeting K-Ras Oncogenes.
Biochemistry, 56, 2017
|
|
5HVV
 
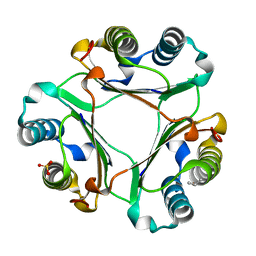 | |
7JX9
 
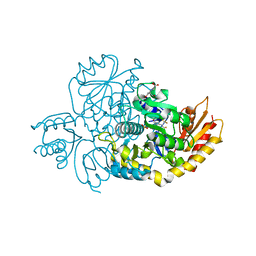 | | The crystal structure of human ornithine aminotransferase with an intermediate bound during inactivation by (1S,3S)-3-amino-4-(hexafluoropropan-2-ylidenyl)-cyclopentane-1-carboxylic acid. | | Descriptor: | (1S,3S,4S)-3-[(E)-({3-hydroxy-2-methyl-5-[(phosphonooxy)methyl]pyridin-4-yl}methylidene)amino]-4-(1,1,3,3,3-pentafluoroprop-1-en-2-yl)cyclopentane-1-carboxylic acid, N-[1,3-dihydroxy-2-(hydroxymethyl)propan-2-yl]glycine, Ornithine aminotransferase, ... | | Authors: | Butrin, A, Beaupre, B, Shen, S, Silverman, R.B, Moran, G, Liu, D. | | Deposit date: | 2020-08-26 | | Release date: | 2021-01-06 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Structural and Kinetic Analyses Reveal the Dual Inhibition Modes of Ornithine Aminotransferase by (1 S ,3 S )-3-Amino-4-(hexafluoropropan-2-ylidenyl)-cyclopentane-1-carboxylic Acid (BCF 3 ).
Acs Chem.Biol., 16, 2021
|
|
1PSP
 
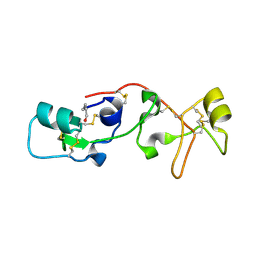 | | PANCREATIC SPASMOLYTIC POLYPEPTIDE: FIRST THREE-DIMENSIONAL STRUCTURE OF A MEMBER OF THE MAMMALIAN TREFOIL FAMILY OF PEPTIDES | | Descriptor: | PANCREATIC SPASMOLYTIC POLYPEPTIDE | | Authors: | Gajhede, M, Petersen, T.N, Henriksen, A, Petersen, J.F.W, Dauter, Z, Wilson, K.S, Thim, L. | | Deposit date: | 1994-01-05 | | Release date: | 1994-04-30 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Pancreatic spasmolytic polypeptide: first three-dimensional structure of a member of the mammalian trefoil family of peptides.
Structure, 1, 1993
|
|
3RIK
 
 | | The acid beta-glucosidase active site exhibits plasticity in binding 3,4,5,6-tetrahydroxyazepane-based inhibitors: implications for pharmacological chaperone design for gaucher disease | | Descriptor: | (3S,4R,5R,6S)-1-(2-hydroxyethyl)azepane-3,4,5,6-tetrol, 2-acetamido-2-deoxy-beta-D-glucopyranose, Glucosylceramidase, ... | | Authors: | Orwig, S.D, Lieberman, R.L. | | Deposit date: | 2011-04-13 | | Release date: | 2012-03-14 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Binding of 3,4,5,6-tetrahydroxyazepanes to the acid-beta-glucosidase active site: implications for pharmacological chaperone design for Gaucher disease
Biochemistry, 50, 2011
|
|
5UMJ
 
 | |
5J5A
 
 | | Trypanosoma brucei methionyl-tRNA synthetase in complex with inhibitor (Chem 70786556) | | Descriptor: | METHIONINE, Methionyl-tRNA synthetase, putative, ... | | Authors: | Barros-Alvarez, X, Koh, C.Y, Hol, W.G.J. | | Deposit date: | 2016-04-01 | | Release date: | 2017-01-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structure-guided design of novel Trypanosoma brucei Methionyl-tRNA synthetase inhibitors.
Eur J Med Chem, 124, 2016
|
|
5I8T
 
 | | Structure of Mouse Acireductone dioxygenase with Ni2+ ion and D-lactic acid in the active site | | Descriptor: | 1,2-dihydroxy-3-keto-5-methylthiopentene dioxygenase, ISOPROPYL ALCOHOL, LACTIC ACID, ... | | Authors: | Deshpande, A.R, Wagenpfeil, K, Pochapsky, T.C, Petsko, G.A, Ringe, D. | | Deposit date: | 2016-02-19 | | Release date: | 2016-03-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.751 Å) | | Cite: | Metal-Dependent Function of a Mammalian Acireductone Dioxygenase.
Biochemistry, 55, 2016
|
|
