4K73
 
 | |
4K74
 
 | |
4K75
 
 | |
4K76
 
 | |
4K77
 
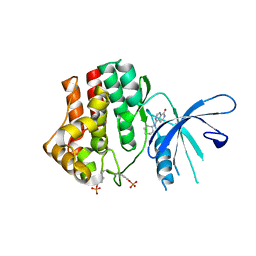 | | JAK1 kinase (JH1 domain) in complex with compound 6 | | Descriptor: | 4-(cyclohexylamino)pyrido[3,4-d]pyrimidin-8(7H)-one, Tyrosine-protein kinase JAK1 | | Authors: | Eigenbrot, C, Shia, S. | | Deposit date: | 2013-04-16 | | Release date: | 2013-10-02 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Design and evaluation of novel 8-oxo-pyridopyrimidine Jak1/2 inhibitors.
Bioorg.Med.Chem.Lett., 23, 2013
|
|
4K78
 
 | |
4K79
 
 | | Recognition of the Thomsen-Friedenreich Antigen by a Lamprey Variable Lymphocyte Receptor | | Descriptor: | Variable lymphocyte receptor, beta-D-galactopyranose-(1-3)-2-acetamido-2-deoxy-alpha-D-galactopyranose | | Authors: | Luo, M, Velikovsky, C.A, Yang, X.B, Mariuzza, R.A. | | Deposit date: | 2013-04-16 | | Release date: | 2013-06-26 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Recognition of the thomsen-friedenreich pancarcinoma carbohydrate antigen by a lamprey variable lymphocyte receptor.
J.Biol.Chem., 288, 2013
|
|
4K7A
 
 | |
4K7B
 
 | | Crystal structure of Extrinsic protein in photosystem II | | Descriptor: | Extrinsic protein in photosystem II, GLYCEROL, SULFATE ION | | Authors: | Nagao, R, Suga, M, Niikura, A, Okumura, A, Koua, F.H.M, Suzuki, T, Tomo, T, Enami, I, Shen, J.R. | | Deposit date: | 2013-04-16 | | Release date: | 2013-09-18 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Crystal Structure of Psb31, a Novel Extrinsic Protein of Photosystem II from a Marine Centric Diatom and Implications for Its Binding and Function
Biochemistry, 52, 2013
|
|
4K7C
 
 | |
4K7D
 
 | | Crystal Structure of Parkin C-terminal RING domains | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase parkin, MALONATE ION, ... | | Authors: | Sauve, V, Trempe, J.-F, Menade, M, Gehring, K. | | Deposit date: | 2013-04-17 | | Release date: | 2013-05-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structure of parkin reveals mechanisms for ubiquitin ligase activation.
Science, 340, 2013
|
|
4K7E
 
 | | Crystal structure of Junin virus nucleoprotein | | Descriptor: | Nucleoprotein | | Authors: | Zhang, Y.J, Li, L, Liu, X, Dong, S.S, Wang, W.M, Huo, T, Rao, Z.H, Yang, C. | | Deposit date: | 2013-04-17 | | Release date: | 2013-08-07 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Junin virus nucleoprotein
J.Gen.Virol., 94, 2013
|
|
4K7F
 
 | | Newly identified epitope V60 from HBV core protein complexed with HLA-A*0201 | | Descriptor: | Beta-2-microglobulin, Core protein, HLA class I histocompatibility antigen, ... | | Authors: | Meng, S.D, Zhang, Y, Wu, Y, Qi, J.X. | | Deposit date: | 2013-04-17 | | Release date: | 2013-06-05 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The L60V variation in hepatitis B virus core protein elicits new epitope-specific cytotoxic T lymphocytes and enhances viral replication.
J.Virol., 87, 2013
|
|
4K7G
 
 | | Crystal structure of a 3-hydroxyproline dehydratse from agrobacterium vitis, target efi-506470, with bound pyrrole 2-carboxylate, ordered active site | | Descriptor: | 3-hydroxyproline dehydratse, ACETATE ION, CITRIC ACID, ... | | Authors: | Vetting, M.W, Toro, R, Bhosle, R, Al Obaidi, N.F, Morisco, L.L, Wasserman, S.R, Sojitra, S, Stead, M, Washington, E, Scott Glenn, A, Chowdhury, S, Evans, B, Hammonds, J, Hillerich, B, Love, J, Seidel, R.D, Imker, H.J, Gerlt, J.A, Almo, S.C, Enzyme Function Initiative (EFI) | | Deposit date: | 2013-04-17 | | Release date: | 2013-05-01 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of a 3-hydroxyproline dehydratse from agrobacterium vitis, target efi-506470, with bound pyrrole 2-carboxylate, ordered active site
To be Published
|
|
4K7H
 
 | | Major capsid protein P1 of the Pseudomonas phage phi6 | | Descriptor: | Major inner protein P1 | | Authors: | Boura, E, Nemecek, D, Plevka, P, Steven, C.A, Hurley, J.H. | | Deposit date: | 2013-04-17 | | Release date: | 2013-08-14 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (3.5964 Å) | | Cite: | Subunit Folds and Maturation Pathway of a dsRNA Virus Capsid.
Structure, 21, 2013
|
|
4K7I
 
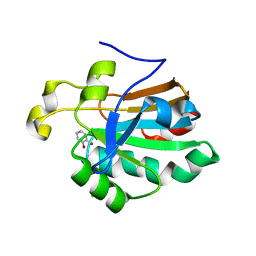 | | HUMAN PEROXIREDOXIN 5 with a fragment | | Descriptor: | CATECHOL, DIMETHYL SULFOXIDE, Peroxiredoxin-5, ... | | Authors: | Guichou, J.F. | | Deposit date: | 2013-04-17 | | Release date: | 2014-04-23 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Comparing Binding Modes of Analogous Fragments Using NMR in Fragment-Based Drug Design: Application to PRDX5
Plos One, 9, 2014
|
|
4K7J
 
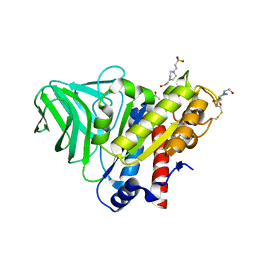 | | Peptidoglycan O-acetylesterase in action, 5 min | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, ACETATE ION, GDSL-like Lipase/Acylhydrolase family protein | | Authors: | Williams, A.H, Gomperts Boneca, I. | | Deposit date: | 2013-04-17 | | Release date: | 2014-09-03 | | Last modified: | 2025-03-26 | | Method: | X-RAY DIFFRACTION (1.968 Å) | | Cite: | Visualization of a substrate-induced productive conformation of the catalytic triad of the Neisseria meningitidis peptidoglycan O-acetylesterase reveals mechanistic conservation in SGNH esterase family members.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4K7K
 
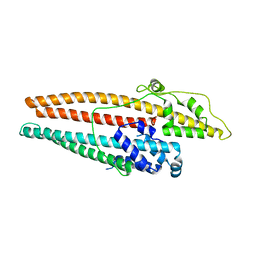 | |
4K7L
 
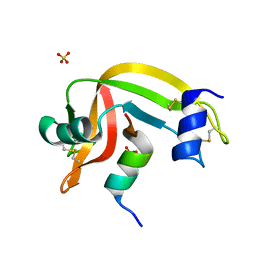 | | Crystal structure of RNase S variant (K7C/Q11C) | | Descriptor: | Ribonuclease pancreatic, SULFATE ION | | Authors: | Genz, M, Straeter, N. | | Deposit date: | 2013-04-17 | | Release date: | 2014-05-14 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Crystal structure of RNase S with a [Hg(Cys2)] metal center in the S-peptide as a template for structure-based design of artificial metalloenzymes using peptide-protein complementation
To be Published
|
|
4K7M
 
 | | Crystal structure of RNase S variant (K7C/Q11C) with bound mercury ions | | Descriptor: | MERCURY (II) ION, Ribonuclease pancreatic, SULFATE ION | | Authors: | Genz, M, Straeter, N. | | Deposit date: | 2013-04-17 | | Release date: | 2013-11-27 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of Apo- and Metalated Thiolate containing RNase S as Structural Basis for the Design of Artificial Metalloenzymes by Peptide- Protein Complementation
Z.Anorg.Allg.Chem., 639, 2013
|
|
4K7N
 
 | | HUMAN PEROXIREDOXIN 5 with a fragment | | Descriptor: | 4-METHYLCATECHOL, Peroxiredoxin-5, mitochondrial | | Authors: | Guichou, J.F. | | Deposit date: | 2013-04-17 | | Release date: | 2014-04-23 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Comparing Binding Modes of Analogous Fragments Using NMR in Fragment-Based Drug Design: Application to PRDX5
Plos One, 9, 2014
|
|
4K7O
 
 | | HUMAN PEROXIREDOXIN 5 with a fragment | | Descriptor: | 4-tert-butylbenzene-1,2-diol, DIMETHYL SULFOXIDE, Peroxiredoxin-5, ... | | Authors: | Guichou, J.F. | | Deposit date: | 2013-04-17 | | Release date: | 2014-04-23 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Comparing Binding Modes of Analogous Fragments Using NMR in Fragment-Based Drug Design: Application to PRDX5
Plos One, 9, 2014
|
|
4K7P
 
 | |
4K7Q
 
 | | Crystal Structure of AcrB Complexed with Linezolid at 3.5 Resolution | | Descriptor: | Acriflavine resistance protein B, N-{[(5S)-3-(3-fluoro-4-morpholin-4-ylphenyl)-2-oxo-1,3-oxazolidin-5-yl]methyl}acetamide | | Authors: | Hung, L.W, Kim, H.B, Murakami, S, Gupta, G, Kim, C.Y, Terwilliger, T.C. | | Deposit date: | 2013-04-17 | | Release date: | 2013-07-03 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Crystal structure of AcrB complexed with linezolid at 3.5 Angstrom resolution.
J.Struct.Funct.Genom., 14, 2013
|
|
4K7R
 
 | | Crystal structures of CusC review conformational changes accompanying folding and transmembrane channel formation | | Descriptor: | (2S)-1-(pentanoyloxy)propan-2-yl hexanoate, Cation efflux system protein CusC | | Authors: | Su, C.-C, Lei, H.-T, Bolla, J.R, Yu, E.W. | | Deposit date: | 2013-04-17 | | Release date: | 2013-10-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.094 Å) | | Cite: | Crystal Structures of CusC Review Conformational Changes Accompanying Folding and Transmembrane Channel Formation.
J.Mol.Biol., 426, 2014
|
|
