5R23
 
 | | PanDDA analysis group deposition -- Endothiapepsin in complex with fragment F2X-Entry F03, DMSO-free | | Descriptor: | Endothiapepsin, N-(3-fluorophenyl)-2-(2-methoxyethoxy)acetamide | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (1.029 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R2F
 
 | | PanDDA analysis group deposition -- Auto-refined data of Endothiapepsin for ground state model 02, DMSO-Free | | Descriptor: | Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (1.119 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R2V
 
 | | PanDDA analysis group deposition -- Auto-refined data of Endothiapepsin for ground state model 19, DMSO-Free | | Descriptor: | Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (1.048 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R3C
 
 | | PanDDA analysis group deposition -- Auto-refined data of Endothiapepsin for ground state model 36, DMSO-Free | | Descriptor: | Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (0.949 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R3Q
 
 | | PanDDA analysis group deposition -- Auto-refined data of Endothiapepsin for ground state model 50, DMSO-Free | | Descriptor: | Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (1.009 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R1W
 
 | | PanDDA analysis group deposition -- Endothiapepsin in complex with fragment F2X-Entry C11, DMSO-free | | Descriptor: | Endothiapepsin, N-ethyl-N-(thiophene-2-carbonyl)-beta-alanine | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (1.077 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R2D
 
 | | PanDDA analysis group deposition -- Endothiapepsin in complex with fragment F2X-Entry H11, DMSO-free | | Descriptor: | 1-cyclopentyl-3-[[(2~{S})-oxolan-2-yl]methyl]urea, Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (0.919 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R2T
 
 | | PanDDA analysis group deposition -- Auto-refined data of Endothiapepsin for ground state model 17, DMSO-Free | | Descriptor: | Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (1.009 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R3B
 
 | | PanDDA analysis group deposition -- Auto-refined data of Endothiapepsin for ground state model 35, DMSO-Free | | Descriptor: | Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (1.17 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
5R3P
 
 | | PanDDA analysis group deposition -- Auto-refined data of Endothiapepsin for ground state model 49, DMSO-Free | | Descriptor: | Endothiapepsin | | Authors: | Wollenhaupt, J, Metz, A, Barthel, T, Lima, G.M.A, Heine, A, Mueller, U, Klebe, G, Weiss, M.S. | | Deposit date: | 2020-02-13 | | Release date: | 2020-06-03 | | Last modified: | 2020-07-08 | | Method: | X-RAY DIFFRACTION (1.077 Å) | | Cite: | F2X-Universal and F2X-Entry: Structurally Diverse Compound Libraries for Crystallographic Fragment Screening.
Structure, 28, 2020
|
|
6LXW
 
 | | Cryo-EM structure of human secretory immunoglobulin A in complex with the N-terminal domain of SpsA | | Descriptor: | Immunoglobulin J chain, Interleukin-2,Immunoglobulin heavy constant alpha 1, Polymeric immunoglobulin receptor, ... | | Authors: | Wang, Y, Wang, G, Li, Y, Xiao, J. | | Deposit date: | 2020-02-12 | | Release date: | 2020-05-27 | | Last modified: | 2020-07-22 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Structural insights into secretory immunoglobulin A and its interaction with a pneumococcal adhesin.
Cell Res., 30, 2020
|
|
6LXK
 
 | | Crystal structure of Z2B3 D102R Fab in complex with influenza virus neuraminidase from A/Serbia/NS-601/2014 (H1N1) | | Descriptor: | CALCIUM ION, Heavy chain of Z2B3-D102R Fab, Light chain of Z2B3-D102R Fab, ... | | Authors: | Jiang, H, Peng, W, Qi, J, Chai, Y, Song, H, Shi, Y, Gao, G.F, Wu, Y. | | Deposit date: | 2020-02-11 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.608 Å) | | Cite: | Structure-Based Modification of an Anti-neuraminidase Human Antibody Restores Protection Efficacy against the Drifted Influenza Virus.
Mbio, 11, 2020
|
|
6LXJ
 
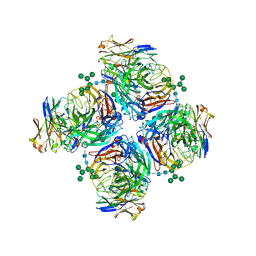 | | Crystal structure of human Z2B3 Fab in complex with influenza virus neuraminidase from A/Anhui/1/2013 (H7N9) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Heavy chain of Z2B3 Fab, ... | | Authors: | Jiang, H, Peng, W, Qi, J, Chai, Y, Song, H, Shi, Y, Gao, G.F, Wu, Y. | | Deposit date: | 2020-02-11 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.903 Å) | | Cite: | Structure-Based Modification of an Anti-neuraminidase Human Antibody Restores Protection Efficacy against the Drifted Influenza Virus.
Mbio, 11, 2020
|
|
6LXI
 
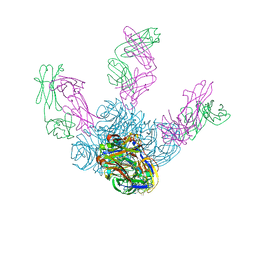 | | Crystal structure of Z2B3 Fab in complex with influenza virus neuraminidase from A/Brevig Mission/1/1918 (H1N1) | | Descriptor: | CALCIUM ION, Heavy chain of Z2B3 Fab, Light chain of Z2B3 Fab, ... | | Authors: | Jiang, H, Peng, W, Qi, J, Chai, Y, Song, H, Shi, Y, Gao, G.F, Wu, Y. | | Deposit date: | 2020-02-11 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure-Based Modification of an Anti-neuraminidase Human Antibody Restores Protection Efficacy against the Drifted Influenza Virus.
Mbio, 11, 2020
|
|
6LXD
 
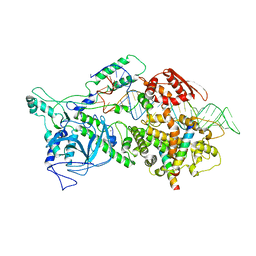 | | Pri-miRNA bound DROSHA-DGCR8 complex | | Descriptor: | Microprocessor complex subunit DGCR8, RNA (102-mer), Ribonuclease 3, ... | | Authors: | Jin, W, Wang, J, Liu, C.P, Wang, H.W, Xu, R.M. | | Deposit date: | 2020-02-10 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Structural Basis for pri-miRNA Recognition by Drosha.
Mol.Cell, 78, 2020
|
|
6LX3
 
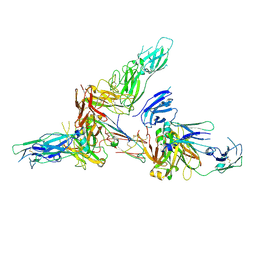 | | Cryo-EM structure of human secretory immunoglobulin A | | Descriptor: | Immunoglobulin J chain, Interleukin-2,Immunoglobulin heavy constant alpha 1, Polymeric immunoglobulin receptor | | Authors: | Wang, Y, Wang, G, Li, Y, Xiao, J. | | Deposit date: | 2020-02-10 | | Release date: | 2020-05-27 | | Last modified: | 2020-07-22 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Structural insights into secretory immunoglobulin A and its interaction with a pneumococcal adhesin.
Cell Res., 30, 2020
|
|
6LXE
 
 | | DROSHA-DGCR8 complex | | Descriptor: | Microprocessor complex subunit DGCR8, Ribonuclease 3, ZINC ION | | Authors: | Jin, W, Wang, J, Liu, C.P, Wang, H.W, Xu, R.M. | | Deposit date: | 2020-02-10 | | Release date: | 2020-04-15 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.2 Å) | | Cite: | Structural Basis for pri-miRNA Recognition by Drosha.
Mol.Cell, 78, 2020
|
|
6Y11
 
 | | Respiratory complex I from Thermus thermophilus | | Descriptor: | DUF3197 domain-containing protein, FE2/S2 (INORGANIC) CLUSTER, FLAVIN MONONUCLEOTIDE, ... | | Authors: | Gutierrez-Fernandez, J, Minhas, G.S, Sazanov, L.A. | | Deposit date: | 2020-02-10 | | Release date: | 2020-09-02 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.109 Å) | | Cite: | Key role of quinone in the mechanism of respiratory complex I.
Nat Commun, 11, 2020
|
|
6VRU
 
 | | PIM-inhibitor complex 1 | | Descriptor: | 3,4-dichloro-2-cyclopropyl-1-[(piperidin-4-yl)methyl]-1H-pyrrolo[2,3-b]pyridine-6-carboxamide, ACETATE ION, IMIDAZOLE, ... | | Authors: | Barberis, C.E, Batchelor, J.D, Mechin, I, Liu, J. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Discovery of SARxxxx92, a pan-PIM kinase inhibitor, efficacious in a KG1 tumor model.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
6VS4
 
 | |
6VRV
 
 | | Discovery of SARxxxx92, a pan-PIM kinase inhibitor, efficacious in a KG1 tumor model | | Descriptor: | 4-chloro-1-{(1S)-1-[(3S)-3-fluoropyrrolidin-3-yl]ethyl}-3-methyl-1H-pyrrolo[2,3-b]pyridine-6-carboxamide, GLYCEROL, Serine/threonine-protein kinase pim-1 | | Authors: | Barberis, C.E, Batchelor, J.D, Liu, J. | | Deposit date: | 2020-02-10 | | Release date: | 2020-11-04 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Discovery of SARxxxx92, a pan-PIM kinase inhibitor, efficacious in a KG1 tumor model.
Bioorg.Med.Chem.Lett., 30, 2020
|
|
6Y0L
 
 | | Crystal structure of human CD23 lectin domain N225D, K229E, S252N, T251N, R253G, S254G mutant | | Descriptor: | GLYCEROL, Low affinity immunoglobulin epsilon Fc receptor membrane-bound form, SULFATE ION | | Authors: | Ilkow, V.F, Davies, A.M, Sutton, B.J, McDonnell, J.M. | | Deposit date: | 2020-02-09 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Reviving lost binding sites: Exploring calcium-binding site transitions between human and murine CD23.
Febs Open Bio, 11, 2021
|
|
6Y0M
 
 | | Crystal structure of human CD23 lectin domain N225D, K229E, S252N, T251N mutant | | Descriptor: | Low affinity immunoglobulin epsilon Fc receptor membrane-bound form | | Authors: | Ilkow, V.F, Davies, A.M, Sutton, B.J, McDonnell, J.M. | | Deposit date: | 2020-02-09 | | Release date: | 2021-06-16 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Reviving lost binding sites: Exploring calcium-binding site transitions between human and murine CD23.
Febs Open Bio, 11, 2021
|
|
6LW5
 
 | | Crystal structure of the human formyl peptide receptor 2 in complex with WKYMVm | | Descriptor: | CHOLESTEROL, Soluble cytochrome b562,N-formyl peptide receptor 2, TRP-LYS-TYR-MET-VAL-QXV | | Authors: | Chen, T, Zong, X, Zhang, H, Wang, M, Zhao, Q, Wu, B. | | Deposit date: | 2020-02-07 | | Release date: | 2020-03-25 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Structural basis of ligand binding modes at the human formyl peptide receptor 2.
Nat Commun, 11, 2020
|
|
6Y09
 
 | |
