9BRD
 
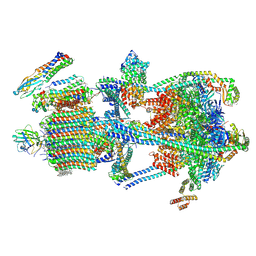 | | Synaptic Vesicle V-ATPase with synaptophysin and SidK, State 3 | | Descriptor: | (7R)-4,7-DIHYDROXY-N,N,N-TRIMETHYL-10-OXO-3,5,9-TRIOXA-4-PHOSPHAHEPTACOSAN-1-AMINIUM 4-OXIDE, 1,2-DIACYL-SN-GLYCERO-3-PHOSPHOCHOLINE, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Coupland, C.E, Rubinstein, J.L. | | Deposit date: | 2024-05-11 | | Release date: | 2024-06-26 | | Last modified: | 2024-07-24 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | High-resolution electron cryomicroscopy of V-ATPase in native synaptic vesicles.
Science, 385, 2024
|
|
8ZCL
 
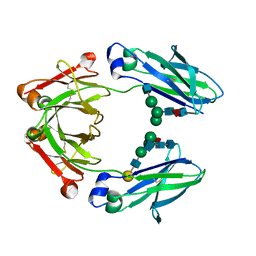 | | Ambient Temperature Crystal Structure of Fc Fragment of Human IgG1 from Biosimilar VEGF-Trap | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin gamma-1 heavy chain, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Destan, E, DeMirci, H. | | Deposit date: | 2024-04-30 | | Release date: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Ambient Temperature Crystal Structure of Fc Fragment of Human IgG1 from Biosimilar VEGF-Trap
To Be Published
|
|
9IIE
 
 | | Cryogenic Temperature Crystal Structure of Fc Fragment of Human IgG1 from Biosimilar VEGF-Trap | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Immunoglobulin gamma-1 heavy chain, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-3)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Destan, E, DeMirci, H. | | Deposit date: | 2024-06-20 | | Release date: | 2024-08-07 | | Method: | X-RAY DIFFRACTION (3.14 Å) | | Cite: | Cryogenic Temperature Crystal Structure of Fc Fragment of Human IgG1 from Biosimilar VEGF-Trap
To Be Published
|
|
9B37
 
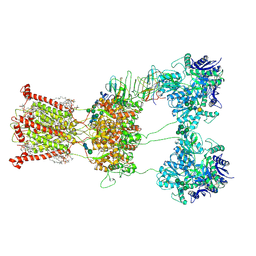 | | Open state of kainate receptor GluK2 in complex with agonist glutamate and positive allosteric modulator BPAM344 bound to one concanavalin A dimer. Composite map. | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Nadezhdin, K.D, Gangwar, S.P, Sobolevsky, A.I. | | Deposit date: | 2024-03-18 | | Release date: | 2024-05-22 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (6.66 Å) | | Cite: | Kainate receptor channel opening and gating mechanism.
Nature, 630, 2024
|
|
9EO0
 
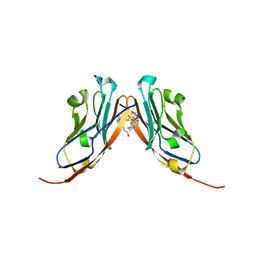 | | Small-Molecule Inhibitors of Programmed Cell Death-1/Programmed Death-Ligand 1 | | Descriptor: | Programmed cell death 1 ligand 1, SULFATE ION, ~{N}-[3-[3-[[5-[(2-hydroxyethylamino)methyl]pyridin-2-yl]carbonylamino]-2-methyl-phenyl]-2-methyl-phenyl]-5-[[3-(methylsulfonylamino)propylamino]methyl]pyridine-2-carboxamide | | Authors: | Plewka, J, Hec, A, Sitar, T, Holak, T. | | Deposit date: | 2024-03-14 | | Release date: | 2024-06-19 | | Last modified: | 2024-07-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Nonsymmetrically Substituted 1,1'-Biphenyl-Based Small Molecule Inhibitors of the PD-1/PD-L1 Interaction.
Acs Med.Chem.Lett., 15, 2024
|
|
6XZA
 
 | | E. coli 70S ribosome in complex with dirithromycin, and deacylated tRNA(iMet) (focused classification). | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Pichkur, E.B, Polikanov, Y.S, Myasnikov, A.G, Konevega, A.L. | | Deposit date: | 2020-02-03 | | Release date: | 2020-11-04 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Insights into the improved macrolide inhibitory activity from the high-resolution cryo-EM structure of dirithromycin bound to the E. coli 70S ribosome.
Rna, 26, 2020
|
|
2DQ5
 
 | | solution structure of the Mid1 B Box2 Chc(D/C)C2H2 Zinc-Binding Domain: insights into an evolutionary conserved ring fold | | Descriptor: | Midline-1, ZINC ION | | Authors: | Massiah, M.A, Matts, J.A.B, Short, K.M, Simmons, B.N, Singireddy, S, Zou, J, Cox, T.C. | | Deposit date: | 2006-05-20 | | Release date: | 2007-04-03 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution Structure of the MID1 B-box2 CHC(D/C)C(2)H(2) Zinc-binding Domain: Insights into an Evolutionarily Conserved RING Fold
J.Mol.Biol., 369, 2007
|
|
3EDK
 
 | | Structural base for cyclodextrin hydrolysis | | Descriptor: | CALCIUM ION, Cyclomaltodextrinase, Cyclooctakis-(1-4)-(alpha-D-glucopyranose), ... | | Authors: | Buedenbender, S, Schulz, G.E. | | Deposit date: | 2008-09-03 | | Release date: | 2009-03-03 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Structural base for enzymatic cyclodextrin hydrolysis
J.Mol.Biol., 385, 2009
|
|
6XKG
 
 | | Crystal structure of 3-O-Sulfotransferase isoform 3 in complex with 8mer oligosaccharide with 6S sulfation | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-6-O-sulfo-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid, 2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-2-O-sulfo-alpha-L-idopyranuronic acid-(1-4)-2-deoxy-6-O-sulfo-2-(sulfoamino)-alpha-D-glucopyranose-(1-4)-beta-D-glucopyranuronic acid, ... | | Authors: | Pedersen, L.C, Liu, J, Wander, R. | | Deposit date: | 2020-06-26 | | Release date: | 2021-06-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Deciphering the substrate recognition mechanisms of the heparan sulfate 3- O -sulfotransferase-3.
Rsc Chem Biol, 2, 2021
|
|
2DW2
 
 | | Crystal structure of VAP2 from Crotalus atrox venom (Form 2-5 crystal) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)][2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Catrocollastatin, ... | | Authors: | Takeda, S, Igarashi, T, Araki, S. | | Deposit date: | 2006-08-02 | | Release date: | 2007-07-10 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structures of catrocollastatin/VAP2B reveal a dynamic, modular architecture of ADAM/adamalysin/reprolysin family proteins
Febs Lett., 581, 2007
|
|
6YSC
 
 | | GLYCOSYLATED KNOB-HOLE/DUMMY FC FRAGMENT | | Descriptor: | Immunoglobulin gamma-1 heavy chain, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kuglstatter, A, Leibrock, L, Benz, J. | | Deposit date: | 2020-04-22 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Format chain exchange (FORCE) for high-throughput generation of bispecific antibodies in combinatorial binder-format matrices.
Nat Commun, 11, 2020
|
|
6Z7P
 
 | | Composite model of the Caulobacter crescentus S-layer bound to the O-antigen of lipopolysaccharide | | Descriptor: | 4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-4-acetamido-4,6-dideoxy-alpha-D-mannopyranose-(1-3)-beta-D-mannopyranose, CALCIUM ION, S-layer protein | | Authors: | Bharat, T.A.M, von Kugelgen, A. | | Deposit date: | 2020-06-01 | | Release date: | 2020-07-15 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (4.8 Å) | | Cite: | In Situ Structure of an Intact Lipopolysaccharide-Bound Bacterial Surface Layer.
Cell, 180, 2020
|
|
6YT7
 
 | | GLYCOSYLATED KNOB/DUMMY-HOLE FC FRAGMENT | | Descriptor: | Ig gamma-1 chain C region, Immunoglobulin gamma-1 heavy chain, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kuglstatter, A, Leibrock, L, Benz, J. | | Deposit date: | 2020-04-24 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Format chain exchange (FORCE) for high-throughput generation of bispecific antibodies in combinatorial binder-format matrices.
Nat Commun, 11, 2020
|
|
6YTB
 
 | | GLYCOSYLATED KNOB/DUMMY-HOLE/DUMMY FC FRAGMENT | | Descriptor: | Immunoglobulin gamma-1 heavy chain, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Kuglstatter, A, Leibrock, L, Benz, J. | | Deposit date: | 2020-04-24 | | Release date: | 2020-10-14 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Format chain exchange (FORCE) for high-throughput generation of bispecific antibodies in combinatorial binder-format matrices.
Nat Commun, 11, 2020
|
|
3GHG
 
 | | Crystal Structure of Human Fibrinogen | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, A knob, B knob, ... | | Authors: | Doolittle, R.F, Kollman, J.M, Sawaya, M.R, Pandi, L, Riley, M. | | Deposit date: | 2009-03-03 | | Release date: | 2009-05-19 | | Last modified: | 2023-05-03 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Crystal structure of human fibrinogen.
Biochemistry, 48, 2009
|
|
6XRT
 
 | |
6XZB
 
 | | E. coli 70S ribosome in complex with dirithromycin, fMet-Phe-tRNA(Phe) and deacylated tRNA(iMet) (focused classification). | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Pichkur, E.B, Polikanov, Y.S, Myasnikov, A.G, Konevega, A.L. | | Deposit date: | 2020-02-03 | | Release date: | 2020-11-04 | | Last modified: | 2024-04-24 | | Method: | ELECTRON MICROSCOPY (2.54 Å) | | Cite: | Insights into the improved macrolide inhibitory activity from the high-resolution cryo-EM structure of dirithromycin bound to the E. coli 70S ribosome.
Rna, 26, 2020
|
|
6YCR
 
 | | Structure of human PD-L1 in complex with inhibitor | | Descriptor: | FFIVIRDRVFR(CCS)G(NH2), Programmed cell death 1 ligand 1 | | Authors: | Magiera-Mularz, K, Grudnik, P, Kuska, K, Holak, T.A, Dubin, G. | | Deposit date: | 2020-03-18 | | Release date: | 2021-02-03 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Macrocyclic Peptide Inhibitor of PD-1/PD-L1 Immune Checkpoint
Adv. Ther., 2020
|
|
2EQD
 
 | | Crystal structure of Cel44A, GH family 44 endoglucanase from Clostridium thermocellum | | Descriptor: | CALCIUM ION, CHLORIDE ION, Endoglucanase, ... | | Authors: | Kitago, Y, Karita, S, Watanabe, N, Sakka, K, Tanaka, I. | | Deposit date: | 2007-03-30 | | Release date: | 2007-09-18 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of Cel44A, a glycoside hydrolase family 44 endoglucanase from Clostridium thermocellum.
J.Biol.Chem., 282, 2007
|
|
3DO3
 
 | | Human 1gG1 Fc fragment, 2.5 Angstrom structure | | Descriptor: | Ig gamma-1 chain C region, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Braden, B.C. | | Deposit date: | 2008-07-03 | | Release date: | 2009-07-21 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Human 1gG1 Fc fragment, 2.5 Angstrom structure.
To be Published
|
|
3EQA
 
 | | Catalytic domain of glucoamylase from aspergillus niger complexed with tris and glycerol | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, GLYCEROL, Glucoamylase, ... | | Authors: | Lee, J, Paetzel, M. | | Deposit date: | 2008-09-30 | | Release date: | 2009-10-13 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of the catalytic domain of glucoamylase from Aspergillus niger.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
2C4A
 
 | | Structure of Neuraminidase Subtype N9 Complexed with 30 MM Sialic Acid (NANA, NEU5AC), Crystal Soaked for 3 Hours at 291 K. | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Rudino-Pinera, E, Tunnah, P, Crennell, S.J, Webster, R.G, Laver, W.G, Garman, E.F. | | Deposit date: | 2005-10-17 | | Release date: | 2007-03-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The Crystal Structure of Influenza Type a Virus Neuraminidase of the N6 Subtype at 1.85 A Resolution
To be Published
|
|
3FJT
 
 | | Crystal structure of a human Fc fragment engineered for extended serum half-life | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, Ig gamma-1 chain C region | | Authors: | Oganesyan, V, Wu, H, Dall'Acqua, W.F. | | Deposit date: | 2008-12-15 | | Release date: | 2009-03-24 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural characterization of a human Fc fragment engineered for extended serum half-life.
Mol.Immunol., 46, 2009
|
|
2D0G
 
 | | Crystal Structure of Thermoactinomyces vulgaris R-47 Alpha-Amylase 1 (TVAI) Mutant D356N/E396Q complexed with P5, a pullulan model oligosaccharide | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, alpha-D-glucopyranose, ... | | Authors: | Abe, A, Yoshida, H, Tonozuka, T, Sakano, Y, Kamitori, S. | | Deposit date: | 2005-08-02 | | Release date: | 2006-07-11 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Complexes of Thermoactinomyces vulgaris R-47 alpha-amylase 1 and pullulan model oligossacharides provide new insight into the mechanism for recognizing substrates with alpha-(1,6) glycosidic linkages
Febs J., 272, 2005
|
|
2END
 
 | | CRYSTAL STRUCTURE OF A PYRIMIDINE DIMER SPECIFIC EXCISION REPAIR ENZYME FROM BACTERIOPHAGE T4: REFINEMENT AT 1.45 ANGSTROMS AND X-RAY ANALYSIS OF THE THREE ACTIVE SITE MUTANTS | | Descriptor: | ENDONUCLEASE V | | Authors: | Vassylyev, D.G, Ariyoshi, M, Matsumoto, O, Katayanagi, K, Ohtsuka, E, Morikawa, K. | | Deposit date: | 1994-08-08 | | Release date: | 1994-10-15 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal structure of a pyrimidine dimer-specific excision repair enzyme from bacteriophage T4: refinement at 1.45 A and X-ray analysis of the three active site mutants.
J.Mol.Biol., 249, 1995
|
|
