6JMZ
 
 | | Structure of H247A mutant open form peptidoglycan peptidase | | Descriptor: | Peptidase M23, ZINC ION | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
8AJN
 
 | | Structure of the human DDB1-DCAF12 complex | | Descriptor: | DDB1- and CUL4-associated factor 12, DNA damage-binding protein 1 | | Authors: | Pla-Prats, C, Cavadini, S, Kempf, G, Thoma, N.H. | | Deposit date: | 2022-07-28 | | Release date: | 2022-11-09 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Recognition of the CCT5 di-Glu degron by CRL4 DCAF12 is dependent on TRiC assembly.
Embo J., 42, 2023
|
|
8AJO
 
 | | Negative-stain electron microscopy structure of DDB1-DCAF12-CCT5 | | Descriptor: | DDB1- and CUL4-associated factor 12, DNA damage-binding protein 1, T-complex protein 1 subunit epsilon | | Authors: | Pla-Prats, C, Cavadini, S, Kempf, G, Thoma, N.H. | | Deposit date: | 2022-07-28 | | Release date: | 2022-11-09 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (30.6 Å) | | Cite: | Recognition of the CCT5 di-Glu degron by CRL4 DCAF12 is dependent on TRiC assembly.
Embo J., 42, 2023
|
|
6MZO
 
 | |
8DFP
 
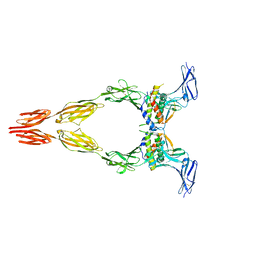 | | Ectodomain of full-length KIT(DupA502,Y503)-SCF dimers | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Isoform 2 of Mast/stem cell growth factor receptor Kit, ... | | Authors: | Bertoletti, N, Krimmer, S.G, Mi, W, Schlessinger, J. | | Deposit date: | 2022-06-22 | | Release date: | 2023-03-29 | | Last modified: | 2023-04-05 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Cryo-EM analyses of KIT and oncogenic mutants reveal structural oncogenic plasticity and a target for therapeutic intervention.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
6N44
 
 | |
1FCG
 
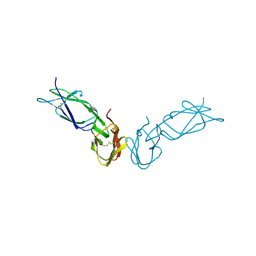 | | ECTODOMAIN OF HUMAN FC GAMMA RECEPTOR, FCGRIIA | | Descriptor: | PROTEIN (FC RECEPTOR FC(GAMMA)RIIA) | | Authors: | Maxwell, K.F, Powell, M.S, Garrett, T.P, Hogarth, P.M. | | Deposit date: | 1999-04-07 | | Release date: | 2000-04-12 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal structure of the human leukocyte Fc receptor, Fc gammaRIIa.
Nat.Struct.Biol., 6, 1999
|
|
6MOZ
 
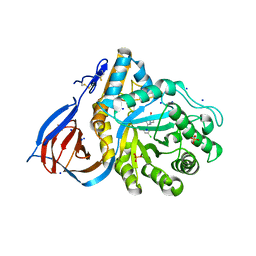 | | Structure of acid-beta-glucosidase in complex with an aromatic pyrrolidine iminosugar inhibitor | | Descriptor: | (2R,3S,4R)-2-{[4-(3,5-dichlorophenyl)-1H-1,2,3-triazol-1-yl]methyl}pyrrolidine-3,4-diol, 2-acetamido-2-deoxy-beta-D-glucopyranose, GLYCEROL, ... | | Authors: | Patterson-Orazem, A.C, Lieberman, R.L. | | Deposit date: | 2018-10-05 | | Release date: | 2019-06-19 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Exploring substituent diversity on pyrrolidine-aryltriazole iminosugars: Structural basis of beta-glucocerebrosidase inhibition.
Bioorg.Chem., 86, 2019
|
|
3NZI
 
 | | Substrate induced remodeling of the active site regulates HtrA1 activity | | Descriptor: | Citrate synthase, Serine protease HTRA1 | | Authors: | Truebestein, L, Tennstaedt, A, Hauske, P, Krojer, T, Kaiser, M, Clausen, T, Ehrmann, M. | | Deposit date: | 2010-07-16 | | Release date: | 2011-02-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Substrate-induced remodeling of the active site regulates human HTRA1 activity.
Nat.Struct.Mol.Biol., 18, 2011
|
|
1FIW
 
 | | THREE-DIMENSIONAL STRUCTURE OF BETA-ACROSIN FROM RAM SPERMATOZOA | | Descriptor: | BETA-ACROSIN HEAVY CHAIN, BETA-ACROSIN LIGHT CHAIN, P-AMINO BENZAMIDINE, ... | | Authors: | Tranter, R, Read, J.A, Jones, R, Brady, R.L. | | Deposit date: | 2000-08-07 | | Release date: | 2000-11-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Effector sites in the three-dimensional structure of mammalian sperm beta-acrosin.
Structure Fold.Des., 8, 2000
|
|
3A4S
 
 | | The crystal structure of the SLD2:Ubc9 complex | | Descriptor: | NFATC2-interacting protein, SUMO-conjugating enzyme UBC9 | | Authors: | Sekiyama, N, Arita, K, Ikeda, Y, Ariyoshi, M, Tochio, H, Saitoh, H, Shirakawa, M. | | Deposit date: | 2009-07-14 | | Release date: | 2010-02-02 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Structural basis for regulation of poly-SUMO chain by a SUMO-like domain of Nip45
Proteins, 78, 2009
|
|
6JMX
 
 | | Structure of open form of peptidoglycan peptidase | | Descriptor: | D(-)-TARTARIC ACID, GLYCEROL, Peptidase M23, ... | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.859 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
6JN1
 
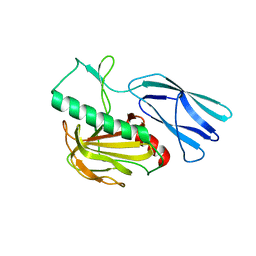 | | Structure of H247A mutant peptidoglycan peptidase complex with penta peptide | | Descriptor: | C0O-DAL-DAL, Peptidase M23, ZINC ION | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.382 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
1FMI
 
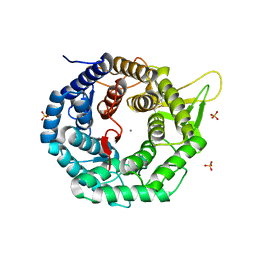 | | CRYSTAL STRUCTURE OF HUMAN CLASS I ALPHA1,2-MANNOSIDASE | | Descriptor: | CALCIUM ION, ENDOPLASMIC RETICULUM ALPHA-MANNOSIDASE I, SULFATE ION | | Authors: | Vallee, F, Karaveg, K, Herscovics, A, Moremen, K.W, Howell, P.L. | | Deposit date: | 2000-08-17 | | Release date: | 2001-01-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural basis for catalysis and inhibition of N-glycan processing class I alpha 1,2-mannosidases.
J.Biol.Chem., 275, 2000
|
|
2BGN
 
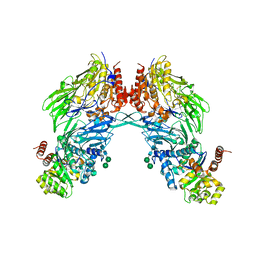 | | HIV-1 Tat protein derived N-terminal nonapeptide Trp2-Tat(1-9) bound to the active site of Dipeptidyl peptidase IV (CD26) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[beta-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Weihofen, W.A, Liu, J, Reutter, W, Saenger, W, Fan, H. | | Deposit date: | 2005-01-03 | | Release date: | 2005-01-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Crystal Structures of HIV-1 Tat-Derived Nonapeptides Tat-(1-9) and Trp2-Tat-(1-9) Bound to the Active Site of Dipeptidyl-Peptidase Iv (Cd26)
J.Biol.Chem., 280, 2005
|
|
1FO3
 
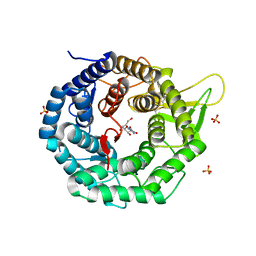 | | CRYSTAL STRUCTURE OF HUMAN CLASS I ALPHA1,2-MANNOSIDASE IN COMPLEX WITH KIFUNENSINE | | Descriptor: | ALPHA1,2-MANNOSIDASE, CALCIUM ION, KIFUNENSINE, ... | | Authors: | Vallee, F, Karaveg, K, Moremen, K.W, Herscovics, A, Howell, P.L. | | Deposit date: | 2000-08-24 | | Release date: | 2001-01-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural basis for catalysis and inhibition of N-glycan processing class I alpha 1,2-mannosidases.
J.Biol.Chem., 275, 2000
|
|
2BLJ
 
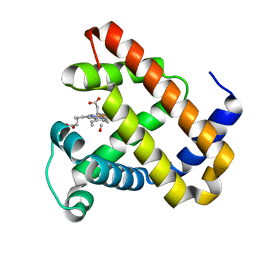 | | Structure of L29W MbCO | | Descriptor: | CARBON MONOXIDE, MYOGLOBIN, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Nienhaus, K, Ostermann, A, Nienhaus, G.U, Parak, F.G, Schmidt, M. | | Deposit date: | 2005-03-04 | | Release date: | 2005-04-06 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Ligand Migration and Protein Fluctuations in Myoglobin Mutant L29W
Biochemistry, 44, 2005
|
|
8ASY
 
 | | SARS-CoV-2 Omicron BA.2.75 RBD in complex with ACE2 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | Authors: | Zhou, D, Huo, J, Ren, J, Stuart, D.I. | | Deposit date: | 2022-08-22 | | Release date: | 2023-01-11 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A delicate balance between antibody evasion and ACE2 affinity for Omicron BA.2.75.
Cell Rep, 42, 2022
|
|
1FIZ
 
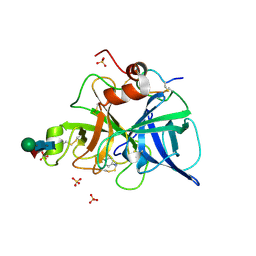 | | THREE DIMENSIONAL STRUCTURE OF BETA-ACROSIN FROM BOAR SPERMATOZOA | | Descriptor: | BETA-ACROSIN HEAVY CHAIN, BETA-ACROSIN LIGHT CHAIN, P-AMINO BENZAMIDINE, ... | | Authors: | Tranter, R, Read, J.A, Jones, R, Brady, R.L. | | Deposit date: | 2000-08-07 | | Release date: | 2000-11-08 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Effector sites in the three-dimensional structure of mammalian sperm beta-acrosin.
Structure Fold.Des., 8, 2000
|
|
1FO0
 
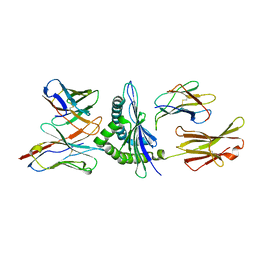 | | MURINE ALLOREACTIVE SCFV TCR-PEPTIDE-MHC CLASS I MOLECULE COMPLEX | | Descriptor: | NATURALLY PROCESSED OCTAPEPTIDE PBM1, PROTEIN (ALLOGENEIC H-2KB MHC CLASS I MOLECULE), PROTEIN (BETA-2 MICROGLOBULIN), ... | | Authors: | Reiser, J.B, Darnault, C, Guimezanes, A, Gregoire, C, Mosser, T, Schmitt-Verhulst, A.-M, Fontecilla-Camps, J.C, Malissen, B, Housset, D, Mazza, G. | | Deposit date: | 2000-08-24 | | Release date: | 2000-10-02 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of a T cell receptor bound to an allogeneic MHC molecule.
Nat.Immunol., 1, 2000
|
|
6MW2
 
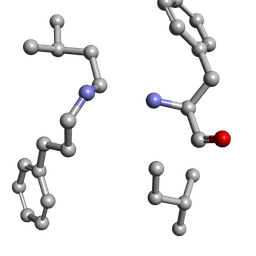 | |
1FSK
 
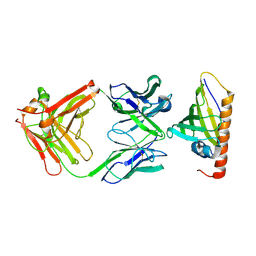 | | COMPLEX FORMATION BETWEEN A FAB FRAGMENT OF A MONOCLONAL IGG ANTIBODY AND THE MAJOR ALLERGEN FROM BIRCH POLLEN BET V 1 | | Descriptor: | ANTIBODY HEAVY CHAIN FAB, IMMUNOGLOBULIN KAPPA LIGHT CHAIN, MAJOR POLLEN ALLERGEN BET V 1-A | | Authors: | Mirza, O, Henriksen, A, Ipsen, H, Larsen, J, Wissenbach, M, Spangfort, M, Gajhede, M. | | Deposit date: | 2000-09-11 | | Release date: | 2000-10-02 | | Last modified: | 2018-03-07 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Dominant epitopes and allergic cross-reactivity: complex formation between a Fab fragment of a monoclonal murine IgG antibody and the major allergen from birch pollen Bet v 1.
J.Immunol., 165, 2000
|
|
1FO2
 
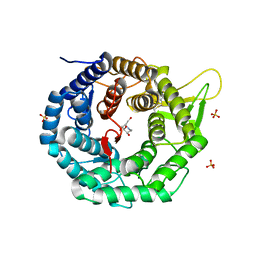 | | CRYSTAL STRUCTURE OF HUMAN CLASS I ALPHA1,2-MANNOSIDASE IN COMPLEX WITH 1-DEOXYMANNOJIRIMYCIN | | Descriptor: | 1-DEOXYMANNOJIRIMYCIN, ALPHA1,2-MANNOSIDASE, CALCIUM ION, ... | | Authors: | Vallee, F, Karaveg, K, Moremen, K.W, Herscovics, A, Howell, P.L. | | Deposit date: | 2000-08-24 | | Release date: | 2001-01-17 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.38 Å) | | Cite: | Structural basis for catalysis and inhibition of N-glycan processing class I alpha 1,2-mannosidases.
J.Biol.Chem., 275, 2000
|
|
1FPU
 
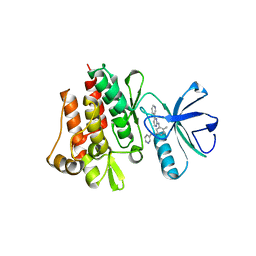 | | CRYSTAL STRUCTURE OF ABL KINASE DOMAIN IN COMPLEX WITH A SMALL MOLECULE INHIBITOR | | Descriptor: | N-[4-METHYL-3-[[4-(3-PYRIDINYL)-2-PYRIMIDINYL]AMINO]PHENYL]-3-PYRIDINECARBOXAMIDE, PROTO-ONCOGENE TYROSINE-PROTEIN KINASE ABL | | Authors: | Schindler, T, Bornmann, W, Pellicena, P, Miller, W.T, Clarkson, B, Kuriyan, J. | | Deposit date: | 2000-08-31 | | Release date: | 2000-09-20 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural mechanism for STI-571 inhibition of abelson tyrosine kinase.
Science, 289, 2000
|
|
6JN0
 
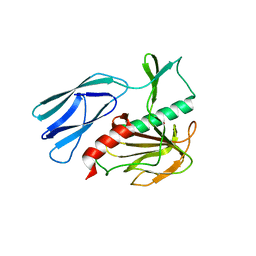 | | Structure of H247A mutant peptidoglycan peptidase complex with tetra-tri peptide | | Descriptor: | C0O-DAL-API, Peptidase M23, ZINC ION | | Authors: | Min, K.J, An, D.R, Yoon, H.J, Suh, S.W, Lee, H.H. | | Deposit date: | 2019-03-13 | | Release date: | 2020-01-15 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.164 Å) | | Cite: | Peptidoglycan reshaping by a noncanonical peptidase for helical cell shape in Campylobacter jejuni.
Nat Commun, 11, 2020
|
|
